
Klebsiella phage KN4-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Przondovirus; Klebsiella virus KN4-1
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
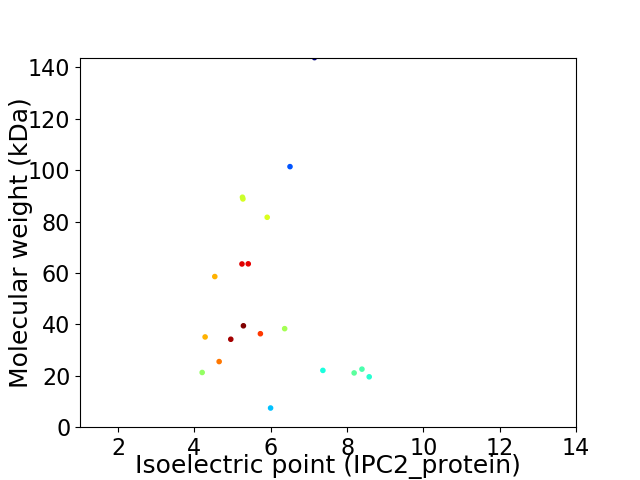
Virtual 2D-PAGE plot for 20 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A3S5IBI0|A0A3S5IBI0_9CAUD Single-stranded DNA-binding protein OS=Klebsiella phage KN4-1 OX=2282631 PE=4 SV=1
MM1 pKa = 7.38NMQDD5 pKa = 4.11AYY7 pKa = 10.62FGSAAEE13 pKa = 3.97LDD15 pKa = 4.2AVNEE19 pKa = 3.86MLAAIGEE26 pKa = 4.7SPVTTLDD33 pKa = 3.33EE34 pKa = 5.18DD35 pKa = 4.25GSADD39 pKa = 3.27VANARR44 pKa = 11.84RR45 pKa = 11.84ILNRR49 pKa = 11.84INRR52 pKa = 11.84QIQSKK57 pKa = 9.61GWAFNINEE65 pKa = 4.55SATLTPDD72 pKa = 2.99ASTGLIPFRR81 pKa = 11.84PAYY84 pKa = 10.44LSILGGQYY92 pKa = 10.17VNRR95 pKa = 11.84GGWVYY100 pKa = 11.2DD101 pKa = 3.54KK102 pKa = 11.07STGTDD107 pKa = 3.14TFSGPITVTMITLQDD122 pKa = 3.63YY123 pKa = 11.57DD124 pKa = 4.19EE125 pKa = 4.72MPEE128 pKa = 5.08CFRR131 pKa = 11.84QWIVTKK137 pKa = 10.66ASRR140 pKa = 11.84QFNSRR145 pKa = 11.84FFGAEE150 pKa = 3.73DD151 pKa = 3.76VEE153 pKa = 4.46NSLAQEE159 pKa = 3.88EE160 pKa = 4.75MEE162 pKa = 5.66ARR164 pKa = 11.84MACNEE169 pKa = 3.84YY170 pKa = 10.91EE171 pKa = 4.14MDD173 pKa = 3.81FGQYY177 pKa = 11.27NMIDD181 pKa = 3.29GDD183 pKa = 4.37SFVGGIIGRR192 pKa = 3.91
MM1 pKa = 7.38NMQDD5 pKa = 4.11AYY7 pKa = 10.62FGSAAEE13 pKa = 3.97LDD15 pKa = 4.2AVNEE19 pKa = 3.86MLAAIGEE26 pKa = 4.7SPVTTLDD33 pKa = 3.33EE34 pKa = 5.18DD35 pKa = 4.25GSADD39 pKa = 3.27VANARR44 pKa = 11.84RR45 pKa = 11.84ILNRR49 pKa = 11.84INRR52 pKa = 11.84QIQSKK57 pKa = 9.61GWAFNINEE65 pKa = 4.55SATLTPDD72 pKa = 2.99ASTGLIPFRR81 pKa = 11.84PAYY84 pKa = 10.44LSILGGQYY92 pKa = 10.17VNRR95 pKa = 11.84GGWVYY100 pKa = 11.2DD101 pKa = 3.54KK102 pKa = 11.07STGTDD107 pKa = 3.14TFSGPITVTMITLQDD122 pKa = 3.63YY123 pKa = 11.57DD124 pKa = 4.19EE125 pKa = 4.72MPEE128 pKa = 5.08CFRR131 pKa = 11.84QWIVTKK137 pKa = 10.66ASRR140 pKa = 11.84QFNSRR145 pKa = 11.84FFGAEE150 pKa = 3.73DD151 pKa = 3.76VEE153 pKa = 4.46NSLAQEE159 pKa = 3.88EE160 pKa = 4.75MEE162 pKa = 5.66ARR164 pKa = 11.84MACNEE169 pKa = 3.84YY170 pKa = 10.91EE171 pKa = 4.14MDD173 pKa = 3.81FGQYY177 pKa = 11.27NMIDD181 pKa = 3.29GDD183 pKa = 4.37SFVGGIIGRR192 pKa = 3.91
Molecular weight: 21.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9WWB4|A0A3Q9WWB4_9CAUD Capsid and scaffold protein OS=Klebsiella phage KN4-1 OX=2282631 PE=4 SV=1
MM1 pKa = 7.79AIALLVEE8 pKa = 4.48SSGVNHH14 pKa = 5.91QPHH17 pKa = 6.94KK18 pKa = 9.79EE19 pKa = 3.64GQMANKK25 pKa = 9.73RR26 pKa = 11.84KK27 pKa = 9.64PIVNGEE33 pKa = 4.28KK34 pKa = 10.12EE35 pKa = 4.13CSKK38 pKa = 10.52CSEE41 pKa = 4.07WKK43 pKa = 9.95PLSEE47 pKa = 4.29YY48 pKa = 10.39GKK50 pKa = 10.62SSGRR54 pKa = 11.84PSGQSQCKK62 pKa = 8.28EE63 pKa = 3.74CRR65 pKa = 11.84KK66 pKa = 9.53SGKK69 pKa = 9.5RR70 pKa = 11.84GKK72 pKa = 7.9PTSAQCRR79 pKa = 11.84SSHH82 pKa = 6.61RR83 pKa = 11.84EE84 pKa = 3.62WILKK88 pKa = 7.13NTYY91 pKa = 10.47GITSEE96 pKa = 4.0EE97 pKa = 4.2FGRR100 pKa = 11.84MLEE103 pKa = 4.25EE104 pKa = 3.59QGGVCRR110 pKa = 11.84ICHH113 pKa = 6.47KK114 pKa = 10.65GPSGRR119 pKa = 11.84FKK121 pKa = 10.69HH122 pKa = 6.21LCVDD126 pKa = 3.88HH127 pKa = 6.79CHH129 pKa = 5.24TTGKK133 pKa = 9.91VRR135 pKa = 11.84GLLCHH140 pKa = 6.98ACNKK144 pKa = 10.39SLGFLEE150 pKa = 5.6DD151 pKa = 3.34DD152 pKa = 3.64PARR155 pKa = 11.84IKK157 pKa = 10.83RR158 pKa = 11.84LIDD161 pKa = 3.38YY162 pKa = 10.44LGGPNEE168 pKa = 4.11PSKK171 pKa = 11.2SVNKK175 pKa = 10.36KK176 pKa = 8.43SS177 pKa = 3.25
MM1 pKa = 7.79AIALLVEE8 pKa = 4.48SSGVNHH14 pKa = 5.91QPHH17 pKa = 6.94KK18 pKa = 9.79EE19 pKa = 3.64GQMANKK25 pKa = 9.73RR26 pKa = 11.84KK27 pKa = 9.64PIVNGEE33 pKa = 4.28KK34 pKa = 10.12EE35 pKa = 4.13CSKK38 pKa = 10.52CSEE41 pKa = 4.07WKK43 pKa = 9.95PLSEE47 pKa = 4.29YY48 pKa = 10.39GKK50 pKa = 10.62SSGRR54 pKa = 11.84PSGQSQCKK62 pKa = 8.28EE63 pKa = 3.74CRR65 pKa = 11.84KK66 pKa = 9.53SGKK69 pKa = 9.5RR70 pKa = 11.84GKK72 pKa = 7.9PTSAQCRR79 pKa = 11.84SSHH82 pKa = 6.61RR83 pKa = 11.84EE84 pKa = 3.62WILKK88 pKa = 7.13NTYY91 pKa = 10.47GITSEE96 pKa = 4.0EE97 pKa = 4.2FGRR100 pKa = 11.84MLEE103 pKa = 4.25EE104 pKa = 3.59QGGVCRR110 pKa = 11.84ICHH113 pKa = 6.47KK114 pKa = 10.65GPSGRR119 pKa = 11.84FKK121 pKa = 10.69HH122 pKa = 6.21LCVDD126 pKa = 3.88HH127 pKa = 6.79CHH129 pKa = 5.24TTGKK133 pKa = 9.91VRR135 pKa = 11.84GLLCHH140 pKa = 6.98ACNKK144 pKa = 10.39SLGFLEE150 pKa = 5.6DD151 pKa = 3.34DD152 pKa = 3.64PARR155 pKa = 11.84IKK157 pKa = 10.83RR158 pKa = 11.84LIDD161 pKa = 3.38YY162 pKa = 10.44LGGPNEE168 pKa = 4.11PSKK171 pKa = 11.2SVNKK175 pKa = 10.36KK176 pKa = 8.43SS177 pKa = 3.25
Molecular weight: 19.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9164 |
69 |
1330 |
458.2 |
50.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.872 ± 0.572 | 1.091 ± 0.235 |
6.427 ± 0.211 | 6.918 ± 0.55 |
3.514 ± 0.178 | 8.468 ± 0.503 |
1.997 ± 0.296 | 4.943 ± 0.261 |
6.198 ± 0.495 | 8.326 ± 0.473 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.543 ± 0.171 | 4.256 ± 0.213 |
3.907 ± 0.166 | 4.114 ± 0.244 |
5.282 ± 0.233 | 6.176 ± 0.551 |
5.227 ± 0.302 | 6.973 ± 0.415 |
1.353 ± 0.247 | 3.416 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
