
candidate division MSBL1 archaeon SCGC-AAA382A03
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
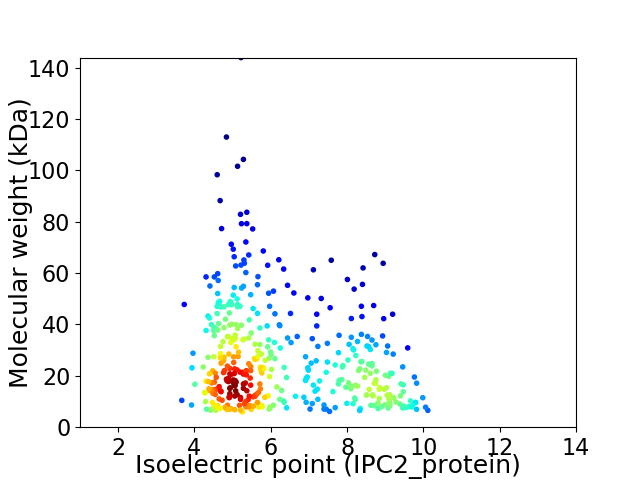
Virtual 2D-PAGE plot for 427 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
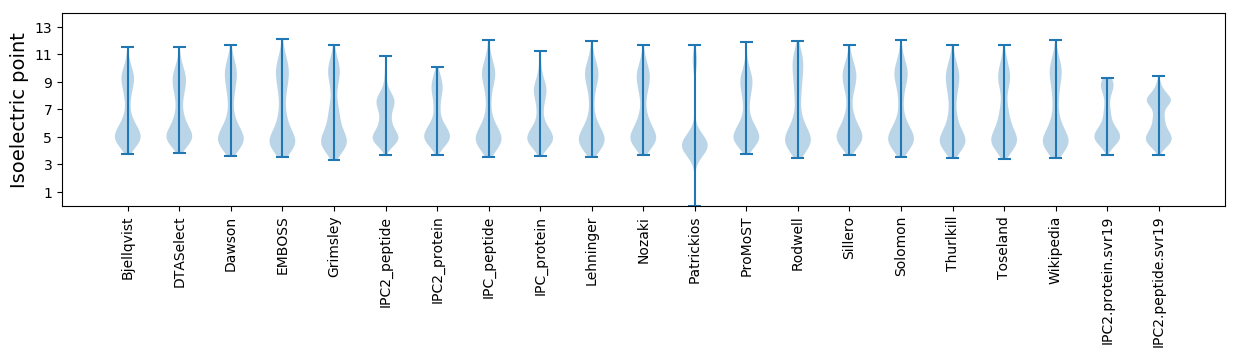
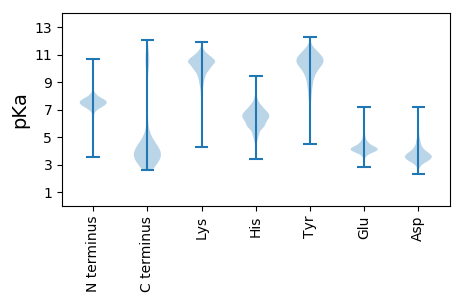
Protein with the lowest isoelectric point:
>tr|A0A133VG05|A0A133VG05_9EURY F420_ligase domain-containing protein OS=candidate division MSBL1 archaeon SCGC-AAA382A03 OX=1698278 GN=AKJ49_00930 PE=4 SV=1
MM1 pKa = 7.42GGTYY5 pKa = 10.64GSLDD9 pKa = 3.7STSGSSPGFSATSEE23 pKa = 4.23TGDD26 pKa = 3.6HH27 pKa = 6.69SSHH30 pKa = 6.97DD31 pKa = 3.72VTVEE35 pKa = 3.78LTVTDD40 pKa = 4.64DD41 pKa = 5.8DD42 pKa = 5.0GATDD46 pKa = 3.62TDD48 pKa = 4.11TATVTVNSQNDD59 pKa = 3.8PPTTSSDD66 pKa = 4.33SITTCEE72 pKa = 4.04NTSGSSDD79 pKa = 3.49VLFNDD84 pKa = 4.25SDD86 pKa = 4.41PEE88 pKa = 4.3GDD90 pKa = 3.43PLVYY94 pKa = 10.77SNVSISTSPSHH105 pKa = 6.57GSASVDD111 pKa = 3.25EE112 pKa = 4.45TAGEE116 pKa = 4.19IVYY119 pKa = 10.69SPNPNYY125 pKa = 10.54NGSDD129 pKa = 3.09SFEE132 pKa = 4.07YY133 pKa = 10.46QISDD137 pKa = 3.56GNGGISTEE145 pKa = 4.41TVSVTVNNVSDD156 pKa = 3.84SPTALFTHH164 pKa = 6.5SSSDD168 pKa = 3.2KK169 pKa = 11.07VLDD172 pKa = 3.53VDD174 pKa = 4.94ASNSSDD180 pKa = 2.94IDD182 pKa = 3.81GSISSYY188 pKa = 6.88EE189 pKa = 4.21WKK191 pKa = 9.82WSSGDD196 pKa = 3.62SFSSGTEE203 pKa = 4.13TDD205 pKa = 2.85SHH207 pKa = 6.55TYY209 pKa = 10.35SSGGEE214 pKa = 3.92YY215 pKa = 9.73TVEE218 pKa = 4.7LRR220 pKa = 11.84VTDD223 pKa = 4.67DD224 pKa = 5.01DD225 pKa = 4.81GATDD229 pKa = 4.1TYY231 pKa = 10.98SQTVTINKK239 pKa = 10.03DD240 pKa = 2.79PDD242 pKa = 3.99APSSPSPSDD251 pKa = 3.41GVKK254 pKa = 10.51LSEE257 pKa = 4.25TSSVTLEE264 pKa = 4.22VTVTDD269 pKa = 4.27PDD271 pKa = 4.27GDD273 pKa = 4.17SMDD276 pKa = 3.42VAFYY280 pKa = 10.98DD281 pKa = 4.49SGGTQIGTTQTGVSDD296 pKa = 3.95GGTTSVSYY304 pKa = 10.65SVSAGNSYY312 pKa = 11.1DD313 pKa = 3.2WYY315 pKa = 11.07AVATDD320 pKa = 4.66DD321 pKa = 5.13EE322 pKa = 5.02GASTQSSTWSFTVKK336 pKa = 10.49PPVDD340 pKa = 3.4EE341 pKa = 4.68SKK343 pKa = 11.32EE344 pKa = 3.94EE345 pKa = 4.1DD346 pKa = 4.88EE347 pKa = 4.61ITQTYY352 pKa = 10.13NLLPILIVLSLSYY365 pKa = 10.76SITLFLSKK373 pKa = 10.22RR374 pKa = 11.84KK375 pKa = 10.04SISTLTHH382 pKa = 4.93KK383 pKa = 10.69RR384 pKa = 11.84IWNVFLLITFLASGILGILLVIRR407 pKa = 11.84INFGFVIPLPFNILFWHH424 pKa = 6.03VEE426 pKa = 3.44AGIAMFVICIFHH438 pKa = 6.57IIEE441 pKa = 4.34RR442 pKa = 11.84FRR444 pKa = 11.84ALLWFLL450 pKa = 4.05
MM1 pKa = 7.42GGTYY5 pKa = 10.64GSLDD9 pKa = 3.7STSGSSPGFSATSEE23 pKa = 4.23TGDD26 pKa = 3.6HH27 pKa = 6.69SSHH30 pKa = 6.97DD31 pKa = 3.72VTVEE35 pKa = 3.78LTVTDD40 pKa = 4.64DD41 pKa = 5.8DD42 pKa = 5.0GATDD46 pKa = 3.62TDD48 pKa = 4.11TATVTVNSQNDD59 pKa = 3.8PPTTSSDD66 pKa = 4.33SITTCEE72 pKa = 4.04NTSGSSDD79 pKa = 3.49VLFNDD84 pKa = 4.25SDD86 pKa = 4.41PEE88 pKa = 4.3GDD90 pKa = 3.43PLVYY94 pKa = 10.77SNVSISTSPSHH105 pKa = 6.57GSASVDD111 pKa = 3.25EE112 pKa = 4.45TAGEE116 pKa = 4.19IVYY119 pKa = 10.69SPNPNYY125 pKa = 10.54NGSDD129 pKa = 3.09SFEE132 pKa = 4.07YY133 pKa = 10.46QISDD137 pKa = 3.56GNGGISTEE145 pKa = 4.41TVSVTVNNVSDD156 pKa = 3.84SPTALFTHH164 pKa = 6.5SSSDD168 pKa = 3.2KK169 pKa = 11.07VLDD172 pKa = 3.53VDD174 pKa = 4.94ASNSSDD180 pKa = 2.94IDD182 pKa = 3.81GSISSYY188 pKa = 6.88EE189 pKa = 4.21WKK191 pKa = 9.82WSSGDD196 pKa = 3.62SFSSGTEE203 pKa = 4.13TDD205 pKa = 2.85SHH207 pKa = 6.55TYY209 pKa = 10.35SSGGEE214 pKa = 3.92YY215 pKa = 9.73TVEE218 pKa = 4.7LRR220 pKa = 11.84VTDD223 pKa = 4.67DD224 pKa = 5.01DD225 pKa = 4.81GATDD229 pKa = 4.1TYY231 pKa = 10.98SQTVTINKK239 pKa = 10.03DD240 pKa = 2.79PDD242 pKa = 3.99APSSPSPSDD251 pKa = 3.41GVKK254 pKa = 10.51LSEE257 pKa = 4.25TSSVTLEE264 pKa = 4.22VTVTDD269 pKa = 4.27PDD271 pKa = 4.27GDD273 pKa = 4.17SMDD276 pKa = 3.42VAFYY280 pKa = 10.98DD281 pKa = 4.49SGGTQIGTTQTGVSDD296 pKa = 3.95GGTTSVSYY304 pKa = 10.65SVSAGNSYY312 pKa = 11.1DD313 pKa = 3.2WYY315 pKa = 11.07AVATDD320 pKa = 4.66DD321 pKa = 5.13EE322 pKa = 5.02GASTQSSTWSFTVKK336 pKa = 10.49PPVDD340 pKa = 3.4EE341 pKa = 4.68SKK343 pKa = 11.32EE344 pKa = 3.94EE345 pKa = 4.1DD346 pKa = 4.88EE347 pKa = 4.61ITQTYY352 pKa = 10.13NLLPILIVLSLSYY365 pKa = 10.76SITLFLSKK373 pKa = 10.22RR374 pKa = 11.84KK375 pKa = 10.04SISTLTHH382 pKa = 4.93KK383 pKa = 10.69RR384 pKa = 11.84IWNVFLLITFLASGILGILLVIRR407 pKa = 11.84INFGFVIPLPFNILFWHH424 pKa = 6.03VEE426 pKa = 3.44AGIAMFVICIFHH438 pKa = 6.57IIEE441 pKa = 4.34RR442 pKa = 11.84FRR444 pKa = 11.84ALLWFLL450 pKa = 4.05
Molecular weight: 47.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133VG00|A0A133VG00_9EURY PKD domain-containing protein OS=candidate division MSBL1 archaeon SCGC-AAA382A03 OX=1698278 GN=AKJ49_00995 PE=4 SV=1
MM1 pKa = 7.55TNPEE5 pKa = 4.07VEE7 pKa = 4.33PTNNRR12 pKa = 11.84AEE14 pKa = 3.97RR15 pKa = 11.84STRR18 pKa = 11.84KK19 pKa = 9.48IVTLRR24 pKa = 11.84KK25 pKa = 9.59IIGTVRR31 pKa = 11.84SEE33 pKa = 3.71RR34 pKa = 11.84GRR36 pKa = 11.84YY37 pKa = 8.56ILEE40 pKa = 4.54TIMTTIEE47 pKa = 3.53TWKK50 pKa = 10.73ARR52 pKa = 11.84GQNPHH57 pKa = 7.17NEE59 pKa = 3.98MQKK62 pKa = 9.91ILRR65 pKa = 11.84NSS67 pKa = 3.16
MM1 pKa = 7.55TNPEE5 pKa = 4.07VEE7 pKa = 4.33PTNNRR12 pKa = 11.84AEE14 pKa = 3.97RR15 pKa = 11.84STRR18 pKa = 11.84KK19 pKa = 9.48IVTLRR24 pKa = 11.84KK25 pKa = 9.59IIGTVRR31 pKa = 11.84SEE33 pKa = 3.71RR34 pKa = 11.84GRR36 pKa = 11.84YY37 pKa = 8.56ILEE40 pKa = 4.54TIMTTIEE47 pKa = 3.53TWKK50 pKa = 10.73ARR52 pKa = 11.84GQNPHH57 pKa = 7.17NEE59 pKa = 3.98MQKK62 pKa = 9.91ILRR65 pKa = 11.84NSS67 pKa = 3.16
Molecular weight: 7.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
103200 |
55 |
1225 |
241.7 |
27.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.797 ± 0.146 | 0.953 ± 0.048 |
5.909 ± 0.106 | 10.261 ± 0.186 |
3.933 ± 0.099 | 6.738 ± 0.131 |
1.668 ± 0.047 | 7.548 ± 0.122 |
8.874 ± 0.174 | 8.82 ± 0.123 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.055 | 4.342 ± 0.073 |
3.944 ± 0.077 | 2.492 ± 0.055 |
5.074 ± 0.118 | 6.283 ± 0.105 |
4.828 ± 0.081 | 6.204 ± 0.096 |
1.052 ± 0.046 | 3.082 ± 0.087 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
