
Human associated gemykibivirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas1
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
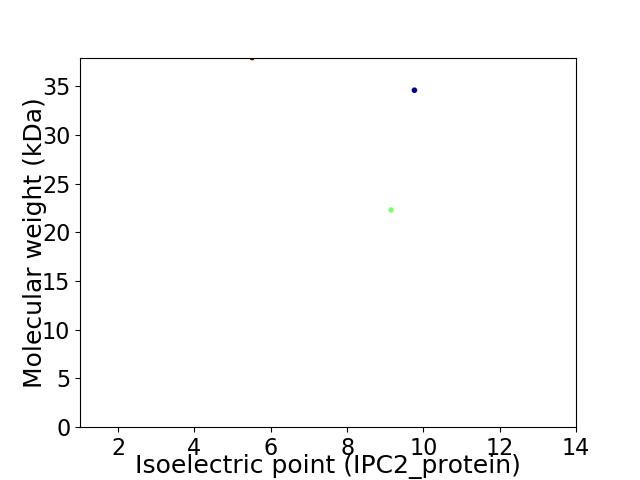
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2C4F9|A0A0S2C4F9_9VIRU Putative ORF3 protein OS=Human associated gemykibivirus 1 OX=2004487 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.7CNNEE5 pKa = 4.03LCLLFDD11 pKa = 5.1FKK13 pKa = 11.05HH14 pKa = 5.68GTPCSPIHH22 pKa = 5.93NVEE25 pKa = 4.0TSILLQWSIILRR37 pKa = 11.84DD38 pKa = 3.74LEE40 pKa = 4.66LNALSAEE47 pKa = 5.07RR48 pKa = 11.84ITQMGGLHH56 pKa = 6.26LHH58 pKa = 6.56AFVDD62 pKa = 4.51FGVKK66 pKa = 9.88YY67 pKa = 8.61RR68 pKa = 11.84TRR70 pKa = 11.84NARR73 pKa = 11.84AFDD76 pKa = 3.62VEE78 pKa = 4.65GFHH81 pKa = 7.21PNVSPSRR88 pKa = 11.84GTPEE92 pKa = 5.32DD93 pKa = 3.67GFDD96 pKa = 3.53YY97 pKa = 10.7AIKK100 pKa = 10.9DD101 pKa = 3.51GDD103 pKa = 4.01VVAGGLEE110 pKa = 4.0RR111 pKa = 11.84PAGSRR116 pKa = 11.84VDD118 pKa = 3.35ATGGVWPEE126 pKa = 3.91IINAKK131 pKa = 10.22DD132 pKa = 3.44EE133 pKa = 4.57SEE135 pKa = 4.12FWSLCEE141 pKa = 3.9SLAPRR146 pKa = 11.84SLVTSFTQLRR156 pKa = 11.84AYY158 pKa = 9.71AAWKK162 pKa = 9.67FPAIRR167 pKa = 11.84EE168 pKa = 4.18PYY170 pKa = 7.05EE171 pKa = 3.87TPEE174 pKa = 4.16GVNIDD179 pKa = 3.83TSWVVEE185 pKa = 3.91LDD187 pKa = 2.93NWVQQNLGRR196 pKa = 11.84APRR199 pKa = 11.84SSAHH203 pKa = 5.92YY204 pKa = 10.1KK205 pKa = 9.61SEE207 pKa = 3.6CRR209 pKa = 11.84LRR211 pKa = 11.84YY212 pKa = 7.15TEE214 pKa = 3.92RR215 pKa = 11.84RR216 pKa = 11.84KK217 pKa = 10.65SLVVYY222 pKa = 10.08GPSRR226 pKa = 11.84MGKK229 pKa = 7.33TIWARR234 pKa = 11.84SLGNHH239 pKa = 7.12AYY241 pKa = 10.41FGGLFSLDD249 pKa = 3.69EE250 pKa = 5.02NIDD253 pKa = 3.52GAEE256 pKa = 3.86YY257 pKa = 10.43AIFDD261 pKa = 4.21DD262 pKa = 4.25FGGIKK267 pKa = 10.32FLPSYY272 pKa = 10.27KK273 pKa = 10.0FWLGHH278 pKa = 4.73QKK280 pKa = 9.95QFYY283 pKa = 8.85VTDD286 pKa = 3.8KK287 pKa = 10.93YY288 pKa = 10.22RR289 pKa = 11.84GKK291 pKa = 10.64RR292 pKa = 11.84LVHH295 pKa = 6.25WARR298 pKa = 11.84PSIWLSNSDD307 pKa = 3.6PRR309 pKa = 11.84DD310 pKa = 3.43EE311 pKa = 5.63LGVDD315 pKa = 4.0TEE317 pKa = 4.54WLNANCDD324 pKa = 3.81FVYY327 pKa = 10.57LDD329 pKa = 4.01SPIVSS334 pKa = 3.68
MM1 pKa = 7.7CNNEE5 pKa = 4.03LCLLFDD11 pKa = 5.1FKK13 pKa = 11.05HH14 pKa = 5.68GTPCSPIHH22 pKa = 5.93NVEE25 pKa = 4.0TSILLQWSIILRR37 pKa = 11.84DD38 pKa = 3.74LEE40 pKa = 4.66LNALSAEE47 pKa = 5.07RR48 pKa = 11.84ITQMGGLHH56 pKa = 6.26LHH58 pKa = 6.56AFVDD62 pKa = 4.51FGVKK66 pKa = 9.88YY67 pKa = 8.61RR68 pKa = 11.84TRR70 pKa = 11.84NARR73 pKa = 11.84AFDD76 pKa = 3.62VEE78 pKa = 4.65GFHH81 pKa = 7.21PNVSPSRR88 pKa = 11.84GTPEE92 pKa = 5.32DD93 pKa = 3.67GFDD96 pKa = 3.53YY97 pKa = 10.7AIKK100 pKa = 10.9DD101 pKa = 3.51GDD103 pKa = 4.01VVAGGLEE110 pKa = 4.0RR111 pKa = 11.84PAGSRR116 pKa = 11.84VDD118 pKa = 3.35ATGGVWPEE126 pKa = 3.91IINAKK131 pKa = 10.22DD132 pKa = 3.44EE133 pKa = 4.57SEE135 pKa = 4.12FWSLCEE141 pKa = 3.9SLAPRR146 pKa = 11.84SLVTSFTQLRR156 pKa = 11.84AYY158 pKa = 9.71AAWKK162 pKa = 9.67FPAIRR167 pKa = 11.84EE168 pKa = 4.18PYY170 pKa = 7.05EE171 pKa = 3.87TPEE174 pKa = 4.16GVNIDD179 pKa = 3.83TSWVVEE185 pKa = 3.91LDD187 pKa = 2.93NWVQQNLGRR196 pKa = 11.84APRR199 pKa = 11.84SSAHH203 pKa = 5.92YY204 pKa = 10.1KK205 pKa = 9.61SEE207 pKa = 3.6CRR209 pKa = 11.84LRR211 pKa = 11.84YY212 pKa = 7.15TEE214 pKa = 3.92RR215 pKa = 11.84RR216 pKa = 11.84KK217 pKa = 10.65SLVVYY222 pKa = 10.08GPSRR226 pKa = 11.84MGKK229 pKa = 7.33TIWARR234 pKa = 11.84SLGNHH239 pKa = 7.12AYY241 pKa = 10.41FGGLFSLDD249 pKa = 3.69EE250 pKa = 5.02NIDD253 pKa = 3.52GAEE256 pKa = 3.86YY257 pKa = 10.43AIFDD261 pKa = 4.21DD262 pKa = 4.25FGGIKK267 pKa = 10.32FLPSYY272 pKa = 10.27KK273 pKa = 10.0FWLGHH278 pKa = 4.73QKK280 pKa = 9.95QFYY283 pKa = 8.85VTDD286 pKa = 3.8KK287 pKa = 10.93YY288 pKa = 10.22RR289 pKa = 11.84GKK291 pKa = 10.64RR292 pKa = 11.84LVHH295 pKa = 6.25WARR298 pKa = 11.84PSIWLSNSDD307 pKa = 3.6PRR309 pKa = 11.84DD310 pKa = 3.43EE311 pKa = 5.63LGVDD315 pKa = 4.0TEE317 pKa = 4.54WLNANCDD324 pKa = 3.81FVYY327 pKa = 10.57LDD329 pKa = 4.01SPIVSS334 pKa = 3.68
Molecular weight: 37.93 kDa
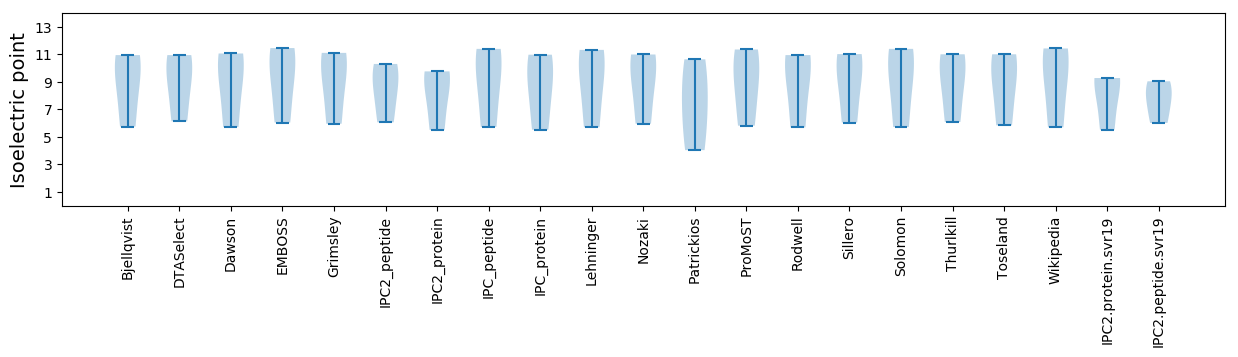
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2C4K3|A0A0S2C4K3_9VIRU Capsid protein OS=Human associated gemykibivirus 1 OX=2004487 GN=Cap PE=4 SV=1
MM1 pKa = 7.1SVARR5 pKa = 11.84RR6 pKa = 11.84SVGRR10 pKa = 11.84TRR12 pKa = 11.84KK13 pKa = 9.36AFRR16 pKa = 11.84RR17 pKa = 11.84GGRR20 pKa = 11.84ATRR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84STRR30 pKa = 11.84FSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84PMTARR46 pKa = 11.84RR47 pKa = 11.84ILNVTSRR54 pKa = 11.84KK55 pKa = 9.84KK56 pKa = 10.06VDD58 pKa = 3.09NMLPIVVDD66 pKa = 3.77EE67 pKa = 4.46EE68 pKa = 4.95SVVTVGPFSSPSPLLCLFVPNARR91 pKa = 11.84DD92 pKa = 3.21TRR94 pKa = 11.84TPITNPAVRR103 pKa = 11.84NSSDD107 pKa = 2.87IFAVGYY113 pKa = 9.27KK114 pKa = 10.08EE115 pKa = 4.36KK116 pKa = 10.86VRR118 pKa = 11.84LDD120 pKa = 3.42VMGGGTFMWRR130 pKa = 11.84RR131 pKa = 11.84IVFMLKK137 pKa = 10.38GDD139 pKa = 3.86DD140 pKa = 3.74LRR142 pKa = 11.84RR143 pKa = 11.84FMDD146 pKa = 3.47SSNSGNVPAQLFDD159 pKa = 3.28QTTEE163 pKa = 4.13GGCRR167 pKa = 11.84RR168 pKa = 11.84VIGPLLGVTNAQEE181 pKa = 4.12EE182 pKa = 4.31LQKK185 pKa = 11.12YY186 pKa = 6.86VFRR189 pKa = 11.84GQEE192 pKa = 3.87DD193 pKa = 4.01VDD195 pKa = 3.48WADD198 pKa = 3.59QFTAPIDD205 pKa = 3.74TRR207 pKa = 11.84RR208 pKa = 11.84VTVKK212 pKa = 9.27YY213 pKa = 10.99DD214 pKa = 2.89KK215 pKa = 10.55MRR217 pKa = 11.84VIRR220 pKa = 11.84PGNDD224 pKa = 2.5TGASRR229 pKa = 11.84LYY231 pKa = 10.58RR232 pKa = 11.84FWHH235 pKa = 6.51PIRR238 pKa = 11.84RR239 pKa = 11.84TISYY243 pKa = 10.58EE244 pKa = 3.91DD245 pKa = 4.05DD246 pKa = 3.97LEE248 pKa = 6.14SDD250 pKa = 4.19VVGDD254 pKa = 4.43RR255 pKa = 11.84PFSTAGLRR263 pKa = 11.84GVGDD267 pKa = 4.13MYY269 pKa = 11.69VMDD272 pKa = 4.89IMAITNLTPDD282 pKa = 4.11APQTSYY288 pKa = 11.1RR289 pKa = 11.84FSPEE293 pKa = 3.23GSFYY297 pKa = 9.4WHH299 pKa = 6.64EE300 pKa = 4.04RR301 pKa = 3.36
MM1 pKa = 7.1SVARR5 pKa = 11.84RR6 pKa = 11.84SVGRR10 pKa = 11.84TRR12 pKa = 11.84KK13 pKa = 9.36AFRR16 pKa = 11.84RR17 pKa = 11.84GGRR20 pKa = 11.84ATRR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84STRR30 pKa = 11.84FSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84PMTARR46 pKa = 11.84RR47 pKa = 11.84ILNVTSRR54 pKa = 11.84KK55 pKa = 9.84KK56 pKa = 10.06VDD58 pKa = 3.09NMLPIVVDD66 pKa = 3.77EE67 pKa = 4.46EE68 pKa = 4.95SVVTVGPFSSPSPLLCLFVPNARR91 pKa = 11.84DD92 pKa = 3.21TRR94 pKa = 11.84TPITNPAVRR103 pKa = 11.84NSSDD107 pKa = 2.87IFAVGYY113 pKa = 9.27KK114 pKa = 10.08EE115 pKa = 4.36KK116 pKa = 10.86VRR118 pKa = 11.84LDD120 pKa = 3.42VMGGGTFMWRR130 pKa = 11.84RR131 pKa = 11.84IVFMLKK137 pKa = 10.38GDD139 pKa = 3.86DD140 pKa = 3.74LRR142 pKa = 11.84RR143 pKa = 11.84FMDD146 pKa = 3.47SSNSGNVPAQLFDD159 pKa = 3.28QTTEE163 pKa = 4.13GGCRR167 pKa = 11.84RR168 pKa = 11.84VIGPLLGVTNAQEE181 pKa = 4.12EE182 pKa = 4.31LQKK185 pKa = 11.12YY186 pKa = 6.86VFRR189 pKa = 11.84GQEE192 pKa = 3.87DD193 pKa = 4.01VDD195 pKa = 3.48WADD198 pKa = 3.59QFTAPIDD205 pKa = 3.74TRR207 pKa = 11.84RR208 pKa = 11.84VTVKK212 pKa = 9.27YY213 pKa = 10.99DD214 pKa = 2.89KK215 pKa = 10.55MRR217 pKa = 11.84VIRR220 pKa = 11.84PGNDD224 pKa = 2.5TGASRR229 pKa = 11.84LYY231 pKa = 10.58RR232 pKa = 11.84FWHH235 pKa = 6.51PIRR238 pKa = 11.84RR239 pKa = 11.84TISYY243 pKa = 10.58EE244 pKa = 3.91DD245 pKa = 4.05DD246 pKa = 3.97LEE248 pKa = 6.14SDD250 pKa = 4.19VVGDD254 pKa = 4.43RR255 pKa = 11.84PFSTAGLRR263 pKa = 11.84GVGDD267 pKa = 4.13MYY269 pKa = 11.69VMDD272 pKa = 4.89IMAITNLTPDD282 pKa = 4.11APQTSYY288 pKa = 11.1RR289 pKa = 11.84FSPEE293 pKa = 3.23GSFYY297 pKa = 9.4WHH299 pKa = 6.64EE300 pKa = 4.04RR301 pKa = 3.36
Molecular weight: 34.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
829 |
194 |
334 |
276.3 |
31.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.669 ± 0.567 | 1.568 ± 0.359 |
5.669 ± 1.095 | 5.428 ± 0.683 |
4.343 ± 0.767 | 7.358 ± 0.222 |
2.533 ± 0.803 | 4.101 ± 0.353 |
3.378 ± 0.241 | 8.323 ± 1.21 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.895 ± 0.957 | 3.86 ± 0.299 |
4.825 ± 0.466 | 2.774 ± 0.509 |
10.736 ± 2.022 | 8.926 ± 1.188 |
5.428 ± 0.757 | 6.634 ± 1.119 |
2.413 ± 0.472 | 3.136 ± 0.351 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
