
Torque teno virus (isolate Human/Ghana/GH1/1996) (TTV)
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae; Torque teno virus; TT virus group 1
Average proteome isoelectric point is 7.81
Get precalculated fractions of proteins
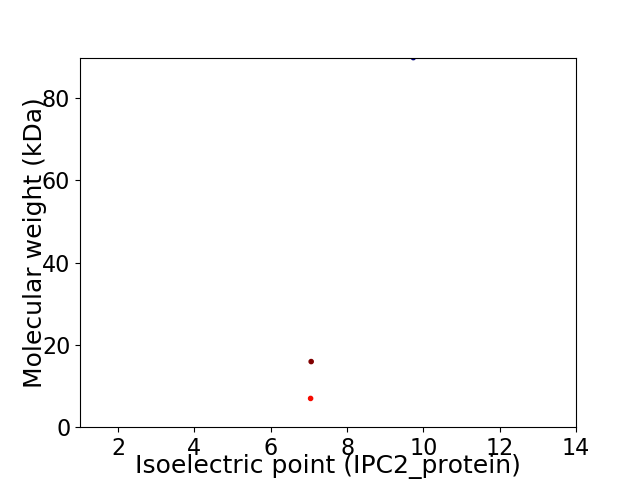
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|Q9WGZ0|CAPSD_TTVVG Probable capsid and replication-associated protein OS=Torque teno virus (isolate Human/Ghana/GH1/1996) OX=487067 GN=ORF1 PE=3 SV=1
MM1 pKa = 7.71FIGRR5 pKa = 11.84HH6 pKa = 4.04YY7 pKa = 10.25RR8 pKa = 11.84KK9 pKa = 9.88KK10 pKa = 10.42RR11 pKa = 11.84ALSLLAVRR19 pKa = 11.84TTQKK23 pKa = 10.64ARR25 pKa = 11.84KK26 pKa = 8.52LLIVMWTPPRR36 pKa = 11.84NDD38 pKa = 3.09QQYY41 pKa = 11.36LNWQWYY47 pKa = 8.81SSVLSSHH54 pKa = 6.96AAMCGCPDD62 pKa = 3.41AVAHH66 pKa = 6.17FNHH69 pKa = 6.91LASVLRR75 pKa = 11.84APQNPPPPGPQRR87 pKa = 11.84NLPLRR92 pKa = 11.84RR93 pKa = 11.84LPALPAAPEE102 pKa = 4.25APGDD106 pKa = 3.71RR107 pKa = 11.84APWPMAGGAEE117 pKa = 4.28GEE119 pKa = 4.24EE120 pKa = 4.7GGAGGDD126 pKa = 3.73ADD128 pKa = 3.99HH129 pKa = 7.08GGAAGGPEE137 pKa = 4.48DD138 pKa = 5.9ADD140 pKa = 4.23LLDD143 pKa = 4.11AVAAAEE149 pKa = 4.19TT150 pKa = 3.88
MM1 pKa = 7.71FIGRR5 pKa = 11.84HH6 pKa = 4.04YY7 pKa = 10.25RR8 pKa = 11.84KK9 pKa = 9.88KK10 pKa = 10.42RR11 pKa = 11.84ALSLLAVRR19 pKa = 11.84TTQKK23 pKa = 10.64ARR25 pKa = 11.84KK26 pKa = 8.52LLIVMWTPPRR36 pKa = 11.84NDD38 pKa = 3.09QQYY41 pKa = 11.36LNWQWYY47 pKa = 8.81SSVLSSHH54 pKa = 6.96AAMCGCPDD62 pKa = 3.41AVAHH66 pKa = 6.17FNHH69 pKa = 6.91LASVLRR75 pKa = 11.84APQNPPPPGPQRR87 pKa = 11.84NLPLRR92 pKa = 11.84RR93 pKa = 11.84LPALPAAPEE102 pKa = 4.25APGDD106 pKa = 3.71RR107 pKa = 11.84APWPMAGGAEE117 pKa = 4.28GEE119 pKa = 4.24EE120 pKa = 4.7GGAGGDD126 pKa = 3.73ADD128 pKa = 3.99HH129 pKa = 7.08GGAAGGPEE137 pKa = 4.48DD138 pKa = 5.9ADD140 pKa = 4.23LLDD143 pKa = 4.11AVAAAEE149 pKa = 4.19TT150 pKa = 3.88
Molecular weight: 15.96 kDa
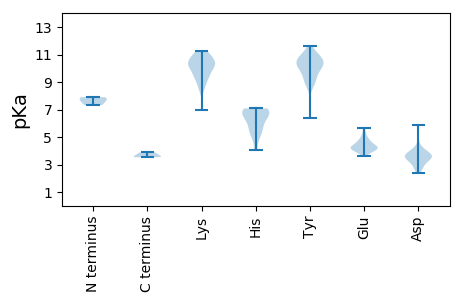
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9WGZ1|YORF3_TTVVG Uncharacterized ORF3 protein OS=Torque teno virus (isolate Human/Ghana/GH1/1996) OX=487067 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.9AYY3 pKa = 9.87GWWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WRR14 pKa = 11.84RR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84PWRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84WRR26 pKa = 11.84TRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84NVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GGRR50 pKa = 11.84WRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.81RR56 pKa = 11.84RR57 pKa = 11.84WKK59 pKa = 9.23RR60 pKa = 11.84KK61 pKa = 6.93GRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84KK66 pKa = 8.81KK67 pKa = 9.54AKK69 pKa = 9.99IIIRR73 pKa = 11.84QWQPNYY79 pKa = 10.06RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84CNIVGYY88 pKa = 10.09IPVLICGEE96 pKa = 4.13NTVSRR101 pKa = 11.84NYY103 pKa = 8.79ATHH106 pKa = 6.96SDD108 pKa = 3.57DD109 pKa = 3.58TNYY112 pKa = 10.32PGPFGGGMTTDD123 pKa = 3.27KK124 pKa = 10.02FTLRR128 pKa = 11.84ILYY131 pKa = 10.27GEE133 pKa = 4.55YY134 pKa = 10.31KK135 pKa = 10.49RR136 pKa = 11.84FMNYY140 pKa = 6.42WTASNEE146 pKa = 3.86DD147 pKa = 3.84LDD149 pKa = 3.9LCRR152 pKa = 11.84YY153 pKa = 10.15LGVNLYY159 pKa = 9.22FFRR162 pKa = 11.84HH163 pKa = 6.39PDD165 pKa = 2.59VDD167 pKa = 5.04FIIKK171 pKa = 9.79INTMPPFLDD180 pKa = 3.76TEE182 pKa = 4.24LTAPSIHH189 pKa = 7.03PGMLALDD196 pKa = 3.83KK197 pKa = 10.25RR198 pKa = 11.84ARR200 pKa = 11.84WIPSLKK206 pKa = 10.18SRR208 pKa = 11.84PGKK211 pKa = 9.5KK212 pKa = 9.19HH213 pKa = 5.44YY214 pKa = 10.48IKK216 pKa = 10.42IRR218 pKa = 11.84VGAPKK223 pKa = 9.99MFTDD227 pKa = 3.05KK228 pKa = 10.35WYY230 pKa = 9.23PQTDD234 pKa = 3.67LCDD237 pKa = 3.56MVLLTVYY244 pKa = 9.17ATAADD249 pKa = 3.61MQYY252 pKa = 11.06PFGSPLTDD260 pKa = 2.83SVVVNFQVLQSMYY273 pKa = 10.54DD274 pKa = 3.49EE275 pKa = 5.65KK276 pKa = 11.09ISILPDD282 pKa = 3.15EE283 pKa = 5.1KK284 pKa = 10.54IQRR287 pKa = 11.84QNLLTSISNYY297 pKa = 9.33IPFYY301 pKa = 10.51NTTQTIAQLKK311 pKa = 8.68PFVDD315 pKa = 3.61AGNAISGTTTTTWGSLLNTTKK336 pKa = 9.72FTTTTTTTYY345 pKa = 9.62TYY347 pKa = 10.57PGTTNTTVTFITANDD362 pKa = 2.63SWYY365 pKa = 10.77RR366 pKa = 11.84GTVYY370 pKa = 10.56NQNIKK375 pKa = 10.74DD376 pKa = 3.92VAKK379 pKa = 10.42KK380 pKa = 10.23AAEE383 pKa = 4.48LYY385 pKa = 10.76SKK387 pKa = 9.45ATKK390 pKa = 10.27AVLGNTFTTEE400 pKa = 4.3DD401 pKa = 3.73YY402 pKa = 10.74TLGYY406 pKa = 10.2HH407 pKa = 6.55GGLYY411 pKa = 10.33SSIWLSPGRR420 pKa = 11.84SYY422 pKa = 11.6FEE424 pKa = 4.11TPGAYY429 pKa = 8.9TDD431 pKa = 3.2IKK433 pKa = 10.91YY434 pKa = 10.91NPFTDD439 pKa = 3.29RR440 pKa = 11.84GEE442 pKa = 4.91GNMLWIDD449 pKa = 3.58WLSKK453 pKa = 11.24KK454 pKa = 10.29NMNYY458 pKa = 10.8DD459 pKa = 3.49KK460 pKa = 11.15VQSKK464 pKa = 10.38CLISDD469 pKa = 4.25LPLWAAAYY477 pKa = 9.96GYY479 pKa = 10.96VEE481 pKa = 5.33FCAKK485 pKa = 9.33STGDD489 pKa = 3.65QNIHH493 pKa = 5.17MNARR497 pKa = 11.84LLIRR501 pKa = 11.84SPFTDD506 pKa = 3.55PQLLVHH512 pKa = 6.69TDD514 pKa = 3.3PTKK517 pKa = 11.11GFVPYY522 pKa = 10.54SLNFGNGKK530 pKa = 8.58MPGGSSNVPIRR541 pKa = 11.84MRR543 pKa = 11.84AKK545 pKa = 9.17WYY547 pKa = 7.43PTLLHH552 pKa = 5.18QQEE555 pKa = 4.42VLEE558 pKa = 4.9ALAQSGPFAYY568 pKa = 10.08HH569 pKa = 6.93ADD571 pKa = 3.38IKK573 pKa = 10.88KK574 pKa = 10.33VSLGMKK580 pKa = 9.93YY581 pKa = 9.58RR582 pKa = 11.84FKK584 pKa = 10.37WIWGGNPVRR593 pKa = 11.84QQVVRR598 pKa = 11.84NPCKK602 pKa = 9.17EE603 pKa = 4.1THH605 pKa = 6.01SSGNRR610 pKa = 11.84VPRR613 pKa = 11.84SLQIVDD619 pKa = 3.88PKK621 pKa = 11.07YY622 pKa = 10.83NSPEE626 pKa = 4.2LTFHH630 pKa = 5.66TWDD633 pKa = 4.02FKK635 pKa = 11.18RR636 pKa = 11.84GLFGPKK642 pKa = 9.78AIQRR646 pKa = 11.84MQQQPTTTDD655 pKa = 2.42IFSAGRR661 pKa = 11.84KK662 pKa = 8.47RR663 pKa = 11.84PRR665 pKa = 11.84RR666 pKa = 11.84DD667 pKa = 2.94TEE669 pKa = 4.53VYY671 pKa = 10.05HH672 pKa = 6.82SSQEE676 pKa = 4.27GEE678 pKa = 4.15QKK680 pKa = 10.66EE681 pKa = 4.53SLLFPPVKK689 pKa = 10.12LLRR692 pKa = 11.84RR693 pKa = 11.84VPPWEE698 pKa = 4.86DD699 pKa = 3.18SQQEE703 pKa = 4.11EE704 pKa = 4.7SGSQSSEE711 pKa = 3.98EE712 pKa = 3.99EE713 pKa = 4.26TQTVSQQLKK722 pKa = 9.41QQLQQQRR729 pKa = 11.84ILGVKK734 pKa = 9.95LRR736 pKa = 11.84LLFNQVQKK744 pKa = 10.59IQQNQDD750 pKa = 2.75INPTLLPRR758 pKa = 11.84GGDD761 pKa = 3.42LASLFQIAPP770 pKa = 3.53
MM1 pKa = 7.9AYY3 pKa = 9.87GWWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WRR14 pKa = 11.84RR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84PWRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84WRR26 pKa = 11.84TRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84NVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GGRR50 pKa = 11.84WRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.81RR56 pKa = 11.84RR57 pKa = 11.84WKK59 pKa = 9.23RR60 pKa = 11.84KK61 pKa = 6.93GRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84KK66 pKa = 8.81KK67 pKa = 9.54AKK69 pKa = 9.99IIIRR73 pKa = 11.84QWQPNYY79 pKa = 10.06RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84CNIVGYY88 pKa = 10.09IPVLICGEE96 pKa = 4.13NTVSRR101 pKa = 11.84NYY103 pKa = 8.79ATHH106 pKa = 6.96SDD108 pKa = 3.57DD109 pKa = 3.58TNYY112 pKa = 10.32PGPFGGGMTTDD123 pKa = 3.27KK124 pKa = 10.02FTLRR128 pKa = 11.84ILYY131 pKa = 10.27GEE133 pKa = 4.55YY134 pKa = 10.31KK135 pKa = 10.49RR136 pKa = 11.84FMNYY140 pKa = 6.42WTASNEE146 pKa = 3.86DD147 pKa = 3.84LDD149 pKa = 3.9LCRR152 pKa = 11.84YY153 pKa = 10.15LGVNLYY159 pKa = 9.22FFRR162 pKa = 11.84HH163 pKa = 6.39PDD165 pKa = 2.59VDD167 pKa = 5.04FIIKK171 pKa = 9.79INTMPPFLDD180 pKa = 3.76TEE182 pKa = 4.24LTAPSIHH189 pKa = 7.03PGMLALDD196 pKa = 3.83KK197 pKa = 10.25RR198 pKa = 11.84ARR200 pKa = 11.84WIPSLKK206 pKa = 10.18SRR208 pKa = 11.84PGKK211 pKa = 9.5KK212 pKa = 9.19HH213 pKa = 5.44YY214 pKa = 10.48IKK216 pKa = 10.42IRR218 pKa = 11.84VGAPKK223 pKa = 9.99MFTDD227 pKa = 3.05KK228 pKa = 10.35WYY230 pKa = 9.23PQTDD234 pKa = 3.67LCDD237 pKa = 3.56MVLLTVYY244 pKa = 9.17ATAADD249 pKa = 3.61MQYY252 pKa = 11.06PFGSPLTDD260 pKa = 2.83SVVVNFQVLQSMYY273 pKa = 10.54DD274 pKa = 3.49EE275 pKa = 5.65KK276 pKa = 11.09ISILPDD282 pKa = 3.15EE283 pKa = 5.1KK284 pKa = 10.54IQRR287 pKa = 11.84QNLLTSISNYY297 pKa = 9.33IPFYY301 pKa = 10.51NTTQTIAQLKK311 pKa = 8.68PFVDD315 pKa = 3.61AGNAISGTTTTTWGSLLNTTKK336 pKa = 9.72FTTTTTTTYY345 pKa = 9.62TYY347 pKa = 10.57PGTTNTTVTFITANDD362 pKa = 2.63SWYY365 pKa = 10.77RR366 pKa = 11.84GTVYY370 pKa = 10.56NQNIKK375 pKa = 10.74DD376 pKa = 3.92VAKK379 pKa = 10.42KK380 pKa = 10.23AAEE383 pKa = 4.48LYY385 pKa = 10.76SKK387 pKa = 9.45ATKK390 pKa = 10.27AVLGNTFTTEE400 pKa = 4.3DD401 pKa = 3.73YY402 pKa = 10.74TLGYY406 pKa = 10.2HH407 pKa = 6.55GGLYY411 pKa = 10.33SSIWLSPGRR420 pKa = 11.84SYY422 pKa = 11.6FEE424 pKa = 4.11TPGAYY429 pKa = 8.9TDD431 pKa = 3.2IKK433 pKa = 10.91YY434 pKa = 10.91NPFTDD439 pKa = 3.29RR440 pKa = 11.84GEE442 pKa = 4.91GNMLWIDD449 pKa = 3.58WLSKK453 pKa = 11.24KK454 pKa = 10.29NMNYY458 pKa = 10.8DD459 pKa = 3.49KK460 pKa = 11.15VQSKK464 pKa = 10.38CLISDD469 pKa = 4.25LPLWAAAYY477 pKa = 9.96GYY479 pKa = 10.96VEE481 pKa = 5.33FCAKK485 pKa = 9.33STGDD489 pKa = 3.65QNIHH493 pKa = 5.17MNARR497 pKa = 11.84LLIRR501 pKa = 11.84SPFTDD506 pKa = 3.55PQLLVHH512 pKa = 6.69TDD514 pKa = 3.3PTKK517 pKa = 11.11GFVPYY522 pKa = 10.54SLNFGNGKK530 pKa = 8.58MPGGSSNVPIRR541 pKa = 11.84MRR543 pKa = 11.84AKK545 pKa = 9.17WYY547 pKa = 7.43PTLLHH552 pKa = 5.18QQEE555 pKa = 4.42VLEE558 pKa = 4.9ALAQSGPFAYY568 pKa = 10.08HH569 pKa = 6.93ADD571 pKa = 3.38IKK573 pKa = 10.88KK574 pKa = 10.33VSLGMKK580 pKa = 9.93YY581 pKa = 9.58RR582 pKa = 11.84FKK584 pKa = 10.37WIWGGNPVRR593 pKa = 11.84QQVVRR598 pKa = 11.84NPCKK602 pKa = 9.17EE603 pKa = 4.1THH605 pKa = 6.01SSGNRR610 pKa = 11.84VPRR613 pKa = 11.84SLQIVDD619 pKa = 3.88PKK621 pKa = 11.07YY622 pKa = 10.83NSPEE626 pKa = 4.2LTFHH630 pKa = 5.66TWDD633 pKa = 4.02FKK635 pKa = 11.18RR636 pKa = 11.84GLFGPKK642 pKa = 9.78AIQRR646 pKa = 11.84MQQQPTTTDD655 pKa = 2.42IFSAGRR661 pKa = 11.84KK662 pKa = 8.47RR663 pKa = 11.84PRR665 pKa = 11.84RR666 pKa = 11.84DD667 pKa = 2.94TEE669 pKa = 4.53VYY671 pKa = 10.05HH672 pKa = 6.82SSQEE676 pKa = 4.27GEE678 pKa = 4.15QKK680 pKa = 10.66EE681 pKa = 4.53SLLFPPVKK689 pKa = 10.12LLRR692 pKa = 11.84RR693 pKa = 11.84VPPWEE698 pKa = 4.86DD699 pKa = 3.18SQQEE703 pKa = 4.11EE704 pKa = 4.7SGSQSSEE711 pKa = 3.98EE712 pKa = 3.99EE713 pKa = 4.26TQTVSQQLKK722 pKa = 9.41QQLQQQRR729 pKa = 11.84ILGVKK734 pKa = 9.95LRR736 pKa = 11.84LLFNQVQKK744 pKa = 10.59IQQNQDD750 pKa = 2.75INPTLLPRR758 pKa = 11.84GGDD761 pKa = 3.42LASLFQIAPP770 pKa = 3.53
Molecular weight: 89.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
977 |
57 |
770 |
325.7 |
37.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.448 ± 3.578 | 1.024 ± 0.235 |
4.913 ± 0.564 | 3.582 ± 0.87 |
3.582 ± 0.787 | 6.96 ± 1.052 |
1.74 ± 0.591 | 4.197 ± 1.662 |
5.732 ± 1.114 | 7.881 ± 1.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.149 ± 0.181 | 4.811 ± 0.908 |
7.472 ± 2.415 | 5.22 ± 0.694 |
9.417 ± 1.09 | 5.425 ± 0.795 |
7.165 ± 2.393 | 4.401 ± 0.685 |
3.071 ± 0.486 | 4.811 ± 1.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
