
Maize yellow dwarf virus RMV
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 8.44
Get precalculated fractions of proteins
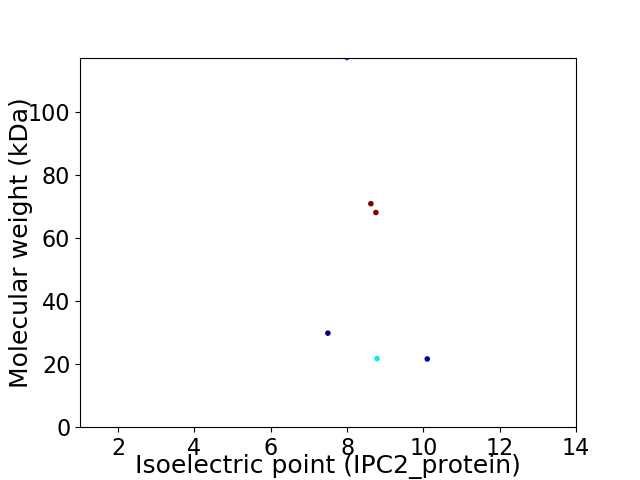
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
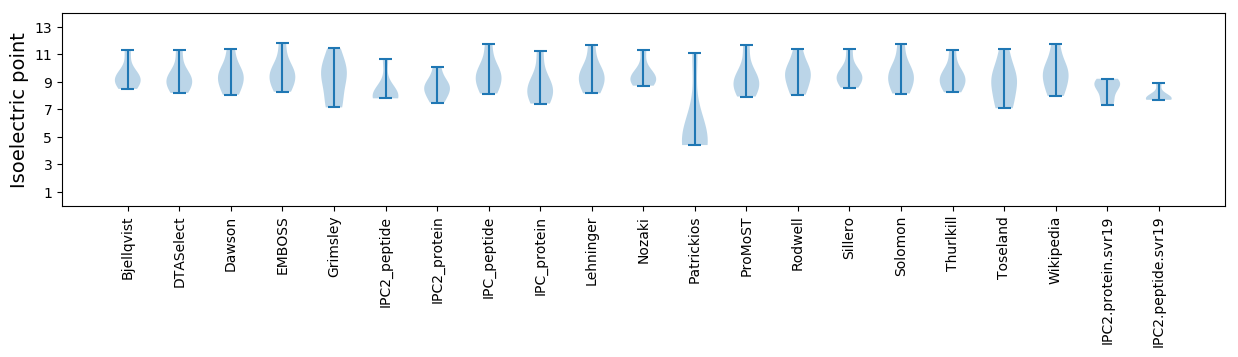
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9U4P5|R9U4P5_9LUTE Movement protein OS=Maize yellow dwarf virus RMV OX=2170101 PE=3 SV=1
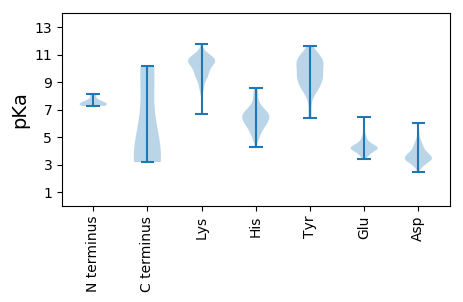
MM1 pKa = 7.46RR2 pKa = 11.84CEE4 pKa = 3.58IHH6 pKa = 7.17FDD8 pKa = 3.35GTLTFAEE15 pKa = 4.63IPRR18 pKa = 11.84DD19 pKa = 3.34LTIRR23 pKa = 11.84LLEE26 pKa = 4.15FQTALRR32 pKa = 11.84TIACCVLNQRR42 pKa = 11.84YY43 pKa = 8.67DD44 pKa = 3.14AEE46 pKa = 4.41TLVRR50 pKa = 11.84SAFTILPLMLRR61 pKa = 11.84EE62 pKa = 5.64LPFTWEE68 pKa = 4.22FPSAVHH74 pKa = 6.69AAFAKK79 pKa = 10.19HH80 pKa = 6.1ALRR83 pKa = 11.84TAGRR87 pKa = 11.84IPPPMEE93 pKa = 5.38LIFHH97 pKa = 7.15GDD99 pKa = 3.69TPKK102 pKa = 10.91HH103 pKa = 5.5MYY105 pKa = 10.23RR106 pKa = 11.84DD107 pKa = 3.39FLYY110 pKa = 10.18RR111 pKa = 11.84VVTRR115 pKa = 11.84DD116 pKa = 2.89IAASLQRR123 pKa = 11.84SHH125 pKa = 6.65EE126 pKa = 4.02NFICGYY132 pKa = 10.44KK133 pKa = 10.13RR134 pKa = 11.84FQLRR138 pKa = 11.84LEE140 pKa = 4.45SVCRR144 pKa = 11.84RR145 pKa = 11.84VEE147 pKa = 3.9LLGDD151 pKa = 3.95TFAPKK156 pKa = 10.1HH157 pKa = 6.0IEE159 pKa = 3.9EE160 pKa = 5.02LSWSSRR166 pKa = 11.84TRR168 pKa = 11.84GGRR171 pKa = 11.84GAGARR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84LPARR182 pKa = 11.84CNLHH186 pKa = 7.22LEE188 pKa = 4.33LLDD191 pKa = 3.45IAALLGCYY199 pKa = 9.79NPGMQIYY206 pKa = 9.5VPDD209 pKa = 4.26VIDD212 pKa = 3.88RR213 pKa = 11.84LSLRR217 pKa = 11.84LYY219 pKa = 10.17NALGPAGFKK228 pKa = 10.7DD229 pKa = 4.42FWRR232 pKa = 11.84VAGYY236 pKa = 10.56DD237 pKa = 3.98HH238 pKa = 7.35LADD241 pKa = 3.52WDD243 pKa = 4.01SDD245 pKa = 3.56IEE247 pKa = 4.12ALSGSVIQKK256 pKa = 9.42EE257 pKa = 4.2LNLLL261 pKa = 3.89
MM1 pKa = 7.46RR2 pKa = 11.84CEE4 pKa = 3.58IHH6 pKa = 7.17FDD8 pKa = 3.35GTLTFAEE15 pKa = 4.63IPRR18 pKa = 11.84DD19 pKa = 3.34LTIRR23 pKa = 11.84LLEE26 pKa = 4.15FQTALRR32 pKa = 11.84TIACCVLNQRR42 pKa = 11.84YY43 pKa = 8.67DD44 pKa = 3.14AEE46 pKa = 4.41TLVRR50 pKa = 11.84SAFTILPLMLRR61 pKa = 11.84EE62 pKa = 5.64LPFTWEE68 pKa = 4.22FPSAVHH74 pKa = 6.69AAFAKK79 pKa = 10.19HH80 pKa = 6.1ALRR83 pKa = 11.84TAGRR87 pKa = 11.84IPPPMEE93 pKa = 5.38LIFHH97 pKa = 7.15GDD99 pKa = 3.69TPKK102 pKa = 10.91HH103 pKa = 5.5MYY105 pKa = 10.23RR106 pKa = 11.84DD107 pKa = 3.39FLYY110 pKa = 10.18RR111 pKa = 11.84VVTRR115 pKa = 11.84DD116 pKa = 2.89IAASLQRR123 pKa = 11.84SHH125 pKa = 6.65EE126 pKa = 4.02NFICGYY132 pKa = 10.44KK133 pKa = 10.13RR134 pKa = 11.84FQLRR138 pKa = 11.84LEE140 pKa = 4.45SVCRR144 pKa = 11.84RR145 pKa = 11.84VEE147 pKa = 3.9LLGDD151 pKa = 3.95TFAPKK156 pKa = 10.1HH157 pKa = 6.0IEE159 pKa = 3.9EE160 pKa = 5.02LSWSSRR166 pKa = 11.84TRR168 pKa = 11.84GGRR171 pKa = 11.84GAGARR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84LPARR182 pKa = 11.84CNLHH186 pKa = 7.22LEE188 pKa = 4.33LLDD191 pKa = 3.45IAALLGCYY199 pKa = 9.79NPGMQIYY206 pKa = 9.5VPDD209 pKa = 4.26VIDD212 pKa = 3.88RR213 pKa = 11.84LSLRR217 pKa = 11.84LYY219 pKa = 10.17NALGPAGFKK228 pKa = 10.7DD229 pKa = 4.42FWRR232 pKa = 11.84VAGYY236 pKa = 10.56DD237 pKa = 3.98HH238 pKa = 7.35LADD241 pKa = 3.52WDD243 pKa = 4.01SDD245 pKa = 3.56IEE247 pKa = 4.12ALSGSVIQKK256 pKa = 9.42EE257 pKa = 4.2LNLLL261 pKa = 3.89
Molecular weight: 29.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9U4P1|R9U4P1_9LUTE PO protein OS=Maize yellow dwarf virus RMV OX=2170101 PE=4 SV=1
MM1 pKa = 7.26NLGGRR6 pKa = 11.84RR7 pKa = 11.84NGRR10 pKa = 11.84RR11 pKa = 11.84GTRR14 pKa = 11.84LRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84VRR20 pKa = 11.84IARR23 pKa = 11.84TTQPMVVVAQTQRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84IRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84PSGDD49 pKa = 2.84TSGGPRR55 pKa = 11.84GRR57 pKa = 11.84GGSRR61 pKa = 11.84EE62 pKa = 3.92TFVFSKK68 pKa = 11.01DD69 pKa = 3.54SIAGSASGKK78 pKa = 8.5LTFGASLSEE87 pKa = 4.16CAAFSGGILKK97 pKa = 10.27AYY99 pKa = 10.17HH100 pKa = 6.69EE101 pKa = 4.34YY102 pKa = 10.8KK103 pKa = 8.01ITKK106 pKa = 9.86VILEE110 pKa = 5.04FISEE114 pKa = 4.35APYY117 pKa = 9.24TAAGSIAYY125 pKa = 9.46EE126 pKa = 3.89LDD128 pKa = 3.22PHH130 pKa = 6.77NKK132 pKa = 9.92LSTLASTINKK142 pKa = 9.47FSIVKK147 pKa = 9.35GGKK150 pKa = 8.72RR151 pKa = 11.84AYY153 pKa = 9.0TSKK156 pKa = 10.86QIGGGVWRR164 pKa = 11.84DD165 pKa = 3.18SSEE168 pKa = 3.95DD169 pKa = 3.37QFAILYY175 pKa = 9.21KK176 pKa = 10.76GSGNSSVAGSFRR188 pKa = 11.84ITMEE192 pKa = 3.75VHH194 pKa = 4.91TQNPKK199 pKa = 10.13
MM1 pKa = 7.26NLGGRR6 pKa = 11.84RR7 pKa = 11.84NGRR10 pKa = 11.84RR11 pKa = 11.84GTRR14 pKa = 11.84LRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84VRR20 pKa = 11.84IARR23 pKa = 11.84TTQPMVVVAQTQRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84IRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84PSGDD49 pKa = 2.84TSGGPRR55 pKa = 11.84GRR57 pKa = 11.84GGSRR61 pKa = 11.84EE62 pKa = 3.92TFVFSKK68 pKa = 11.01DD69 pKa = 3.54SIAGSASGKK78 pKa = 8.5LTFGASLSEE87 pKa = 4.16CAAFSGGILKK97 pKa = 10.27AYY99 pKa = 10.17HH100 pKa = 6.69EE101 pKa = 4.34YY102 pKa = 10.8KK103 pKa = 8.01ITKK106 pKa = 9.86VILEE110 pKa = 5.04FISEE114 pKa = 4.35APYY117 pKa = 9.24TAAGSIAYY125 pKa = 9.46EE126 pKa = 3.89LDD128 pKa = 3.22PHH130 pKa = 6.77NKK132 pKa = 9.92LSTLASTINKK142 pKa = 9.47FSIVKK147 pKa = 9.35GGKK150 pKa = 8.72RR151 pKa = 11.84AYY153 pKa = 9.0TSKK156 pKa = 10.86QIGGGVWRR164 pKa = 11.84DD165 pKa = 3.18SSEE168 pKa = 3.95DD169 pKa = 3.37QFAILYY175 pKa = 9.21KK176 pKa = 10.76GSGNSSVAGSFRR188 pKa = 11.84ITMEE192 pKa = 3.75VHH194 pKa = 4.91TQNPKK199 pKa = 10.13
Molecular weight: 21.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2964 |
193 |
1045 |
494.0 |
55.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.018 ± 0.437 | 1.754 ± 0.323 |
4.285 ± 0.401 | 5.364 ± 0.371 |
3.745 ± 0.299 | 7.085 ± 0.833 |
1.923 ± 0.269 | 4.791 ± 0.341 |
5.128 ± 0.548 | 9.615 ± 1.126 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.518 ± 0.126 | 3.88 ± 0.525 |
5.398 ± 0.633 | 3.981 ± 0.474 |
7.355 ± 1.007 | 9.717 ± 0.716 |
6.377 ± 0.519 | 6.174 ± 0.414 |
1.687 ± 0.214 | 3.171 ± 0.189 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
