
Klebsiella phage KN3-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Przondovirus; Klebsiella virus KN3-1
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
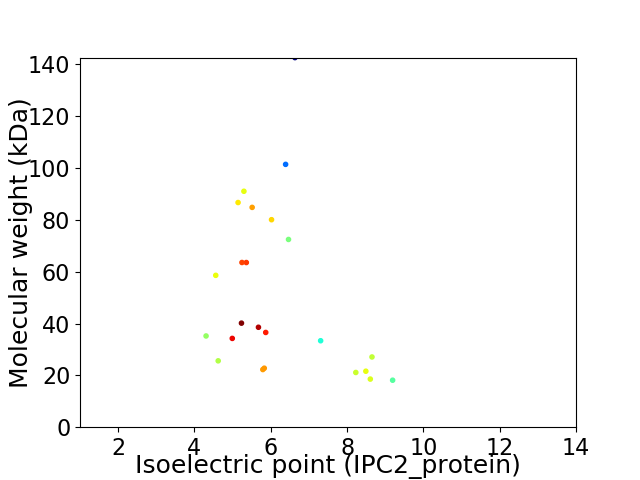
Virtual 2D-PAGE plot for 24 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9WWY9|A0A3Q9WWY9_9CAUD DNA-directed RNA polymerase OS=Klebsiella phage KN3-1 OX=2282630 PE=3 SV=1
MM1 pKa = 7.69AGEE4 pKa = 4.42SNADD8 pKa = 3.27VYY10 pKa = 11.74ASFGVNSAVLTGSTPEE26 pKa = 3.81EE27 pKa = 4.08HH28 pKa = 6.29QEE30 pKa = 3.72NMLALDD36 pKa = 3.66VAARR40 pKa = 11.84DD41 pKa = 3.53GDD43 pKa = 4.19DD44 pKa = 5.06AIEE47 pKa = 4.63LNTNSDD53 pKa = 3.81DD54 pKa = 4.22PYY56 pKa = 11.43GSDD59 pKa = 3.19VDD61 pKa = 3.98PFGEE65 pKa = 4.26PEE67 pKa = 3.93EE68 pKa = 5.05GRR70 pKa = 11.84MQVRR74 pKa = 11.84ISADD78 pKa = 3.11GDD80 pKa = 3.73DD81 pKa = 4.28PEE83 pKa = 4.79EE84 pKa = 3.96EE85 pKa = 4.18VVEE88 pKa = 4.41EE89 pKa = 4.5EE90 pKa = 4.22EE91 pKa = 4.24QQGDD95 pKa = 3.71EE96 pKa = 4.16EE97 pKa = 4.7GQLEE101 pKa = 4.4EE102 pKa = 4.2VTEE105 pKa = 4.18EE106 pKa = 4.44GEE108 pKa = 4.11PEE110 pKa = 3.89EE111 pKa = 4.3FKK113 pKa = 11.04PIGEE117 pKa = 4.36TPADD121 pKa = 3.26ISEE124 pKa = 4.49ASQQLEE130 pKa = 3.83EE131 pKa = 4.93HH132 pKa = 6.15EE133 pKa = 5.18AGFNDD138 pKa = 3.81MVATAIEE145 pKa = 4.42RR146 pKa = 11.84GLSQDD151 pKa = 2.65AVTRR155 pKa = 11.84IQQEE159 pKa = 4.18YY160 pKa = 9.09QNEE163 pKa = 3.96DD164 pKa = 3.44RR165 pKa = 11.84LSDD168 pKa = 3.51EE169 pKa = 4.57SYY171 pKa = 11.21KK172 pKa = 10.72EE173 pKa = 3.78LAEE176 pKa = 4.32AGYY179 pKa = 11.04SKK181 pKa = 11.26AFVDD185 pKa = 4.66AYY187 pKa = 10.18IRR189 pKa = 11.84GQEE192 pKa = 4.01ALVNQYY198 pKa = 8.39VEE200 pKa = 4.13KK201 pKa = 11.31VMDD204 pKa = 3.92FVGGRR209 pKa = 11.84EE210 pKa = 3.86RR211 pKa = 11.84FQQVYY216 pKa = 7.32THH218 pKa = 6.02MQTNNPEE225 pKa = 3.88GAEE228 pKa = 3.93ALIKK232 pKa = 10.63AFEE235 pKa = 4.23SRR237 pKa = 11.84DD238 pKa = 3.48VATMKK243 pKa = 10.36TILNLAGQSRR253 pKa = 11.84DD254 pKa = 3.16KK255 pKa = 11.32TFGKK259 pKa = 10.22KK260 pKa = 9.91AEE262 pKa = 3.85RR263 pKa = 11.84SIAKK267 pKa = 9.62RR268 pKa = 11.84ATPAKK273 pKa = 9.25PVARR277 pKa = 11.84KK278 pKa = 10.1AEE280 pKa = 4.23GFEE283 pKa = 4.58SQAEE287 pKa = 4.38MIKK290 pKa = 10.76AMSDD294 pKa = 2.8PRR296 pKa = 11.84YY297 pKa = 9.04RR298 pKa = 11.84TDD300 pKa = 2.52SKK302 pKa = 11.03YY303 pKa = 10.48RR304 pKa = 11.84RR305 pKa = 11.84EE306 pKa = 4.17VEE308 pKa = 4.0QKK310 pKa = 11.01VIDD313 pKa = 3.98SKK315 pKa = 11.46FF316 pKa = 3.06
MM1 pKa = 7.69AGEE4 pKa = 4.42SNADD8 pKa = 3.27VYY10 pKa = 11.74ASFGVNSAVLTGSTPEE26 pKa = 3.81EE27 pKa = 4.08HH28 pKa = 6.29QEE30 pKa = 3.72NMLALDD36 pKa = 3.66VAARR40 pKa = 11.84DD41 pKa = 3.53GDD43 pKa = 4.19DD44 pKa = 5.06AIEE47 pKa = 4.63LNTNSDD53 pKa = 3.81DD54 pKa = 4.22PYY56 pKa = 11.43GSDD59 pKa = 3.19VDD61 pKa = 3.98PFGEE65 pKa = 4.26PEE67 pKa = 3.93EE68 pKa = 5.05GRR70 pKa = 11.84MQVRR74 pKa = 11.84ISADD78 pKa = 3.11GDD80 pKa = 3.73DD81 pKa = 4.28PEE83 pKa = 4.79EE84 pKa = 3.96EE85 pKa = 4.18VVEE88 pKa = 4.41EE89 pKa = 4.5EE90 pKa = 4.22EE91 pKa = 4.24QQGDD95 pKa = 3.71EE96 pKa = 4.16EE97 pKa = 4.7GQLEE101 pKa = 4.4EE102 pKa = 4.2VTEE105 pKa = 4.18EE106 pKa = 4.44GEE108 pKa = 4.11PEE110 pKa = 3.89EE111 pKa = 4.3FKK113 pKa = 11.04PIGEE117 pKa = 4.36TPADD121 pKa = 3.26ISEE124 pKa = 4.49ASQQLEE130 pKa = 3.83EE131 pKa = 4.93HH132 pKa = 6.15EE133 pKa = 5.18AGFNDD138 pKa = 3.81MVATAIEE145 pKa = 4.42RR146 pKa = 11.84GLSQDD151 pKa = 2.65AVTRR155 pKa = 11.84IQQEE159 pKa = 4.18YY160 pKa = 9.09QNEE163 pKa = 3.96DD164 pKa = 3.44RR165 pKa = 11.84LSDD168 pKa = 3.51EE169 pKa = 4.57SYY171 pKa = 11.21KK172 pKa = 10.72EE173 pKa = 3.78LAEE176 pKa = 4.32AGYY179 pKa = 11.04SKK181 pKa = 11.26AFVDD185 pKa = 4.66AYY187 pKa = 10.18IRR189 pKa = 11.84GQEE192 pKa = 4.01ALVNQYY198 pKa = 8.39VEE200 pKa = 4.13KK201 pKa = 11.31VMDD204 pKa = 3.92FVGGRR209 pKa = 11.84EE210 pKa = 3.86RR211 pKa = 11.84FQQVYY216 pKa = 7.32THH218 pKa = 6.02MQTNNPEE225 pKa = 3.88GAEE228 pKa = 3.93ALIKK232 pKa = 10.63AFEE235 pKa = 4.23SRR237 pKa = 11.84DD238 pKa = 3.48VATMKK243 pKa = 10.36TILNLAGQSRR253 pKa = 11.84DD254 pKa = 3.16KK255 pKa = 11.32TFGKK259 pKa = 10.22KK260 pKa = 9.91AEE262 pKa = 3.85RR263 pKa = 11.84SIAKK267 pKa = 9.62RR268 pKa = 11.84ATPAKK273 pKa = 9.25PVARR277 pKa = 11.84KK278 pKa = 10.1AEE280 pKa = 4.23GFEE283 pKa = 4.58SQAEE287 pKa = 4.38MIKK290 pKa = 10.76AMSDD294 pKa = 2.8PRR296 pKa = 11.84YY297 pKa = 9.04RR298 pKa = 11.84TDD300 pKa = 2.52SKK302 pKa = 11.03YY303 pKa = 10.48RR304 pKa = 11.84RR305 pKa = 11.84EE306 pKa = 4.17VEE308 pKa = 4.0QKK310 pKa = 11.01VIDD313 pKa = 3.98SKK315 pKa = 11.46FF316 pKa = 3.06
Molecular weight: 35.19 kDa
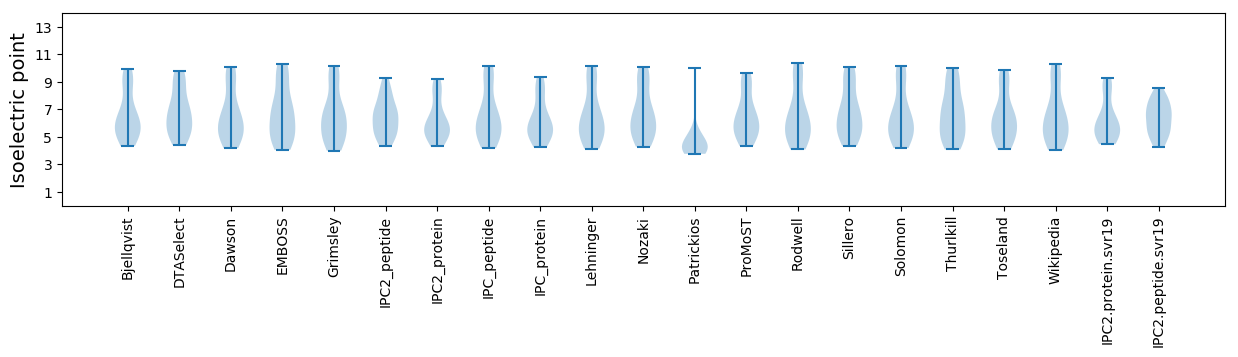
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9WSD5|A0A3Q9WSD5_9CAUD Endolysin OS=Klebsiella phage KN3-1 OX=2282630 PE=3 SV=1
MM1 pKa = 7.9PYY3 pKa = 10.3SSSNHH8 pKa = 5.51LSYY11 pKa = 11.18VPSSMSFCLNSRR23 pKa = 11.84GCCRR27 pKa = 11.84KK28 pKa = 10.02RR29 pKa = 11.84IPLAIPKK36 pKa = 8.31TSSMVSSTDD45 pKa = 2.99SSSIAMPTRR54 pKa = 11.84SPCPLANPRR63 pKa = 11.84ACWRR67 pKa = 11.84TNVDD71 pKa = 4.43LPIPDD76 pKa = 4.8PEE78 pKa = 4.33VTMMTSPPRR87 pKa = 11.84APKK90 pKa = 10.02VRR92 pKa = 11.84SFSPGHH98 pKa = 5.51PEE100 pKa = 3.28KK101 pKa = 10.48RR102 pKa = 11.84YY103 pKa = 7.6PTLCSLVMASRR114 pKa = 11.84TLSFMDD120 pKa = 4.14IAPITPSGTQGAAFHH135 pKa = 6.44IWSSISLPLPLSKK148 pKa = 10.63HH149 pKa = 5.34SLASFSVSTATWTLPGGRR167 pKa = 11.84TT168 pKa = 3.32
MM1 pKa = 7.9PYY3 pKa = 10.3SSSNHH8 pKa = 5.51LSYY11 pKa = 11.18VPSSMSFCLNSRR23 pKa = 11.84GCCRR27 pKa = 11.84KK28 pKa = 10.02RR29 pKa = 11.84IPLAIPKK36 pKa = 8.31TSSMVSSTDD45 pKa = 2.99SSSIAMPTRR54 pKa = 11.84SPCPLANPRR63 pKa = 11.84ACWRR67 pKa = 11.84TNVDD71 pKa = 4.43LPIPDD76 pKa = 4.8PEE78 pKa = 4.33VTMMTSPPRR87 pKa = 11.84APKK90 pKa = 10.02VRR92 pKa = 11.84SFSPGHH98 pKa = 5.51PEE100 pKa = 3.28KK101 pKa = 10.48RR102 pKa = 11.84YY103 pKa = 7.6PTLCSLVMASRR114 pKa = 11.84TLSFMDD120 pKa = 4.14IAPITPSGTQGAAFHH135 pKa = 6.44IWSSISLPLPLSKK148 pKa = 10.63HH149 pKa = 5.34SLASFSVSTATWTLPGGRR167 pKa = 11.84TT168 pKa = 3.32
Molecular weight: 18.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11193 |
167 |
1321 |
466.4 |
51.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.702 ± 0.624 | 1.126 ± 0.214 |
6.352 ± 0.219 | 6.522 ± 0.511 |
3.565 ± 0.207 | 8.184 ± 0.414 |
2.073 ± 0.275 | 4.664 ± 0.18 |
5.754 ± 0.45 | 8.684 ± 0.653 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.51 ± 0.197 | 4.181 ± 0.229 |
3.877 ± 0.263 | 4.494 ± 0.332 |
5.557 ± 0.268 | 6.057 ± 0.364 |
5.87 ± 0.332 | 7.281 ± 0.508 |
1.242 ± 0.18 | 3.306 ± 0.275 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
