
Hubei sobemo-like virus 49
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
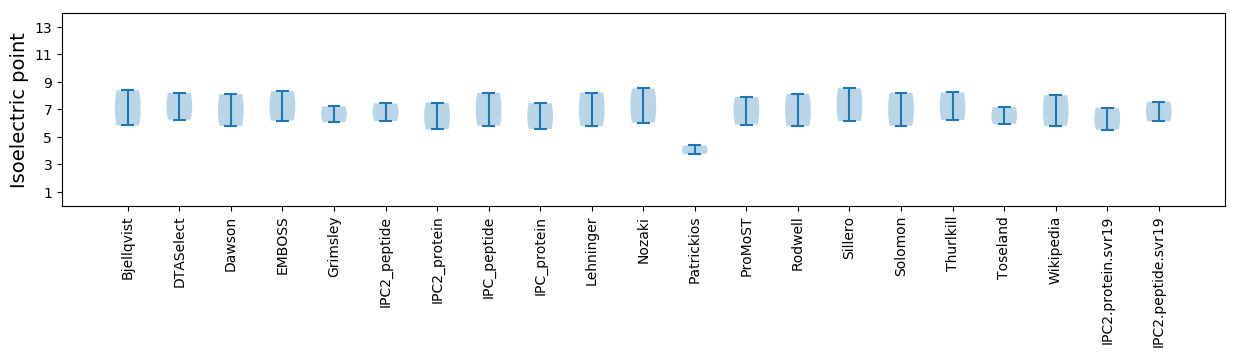
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEH2|A0A1L3KEH2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 49 OX=1923237 PE=4 SV=1
MM1 pKa = 7.3TSSPGYY7 pKa = 9.38PYY9 pKa = 9.94MRR11 pKa = 11.84EE12 pKa = 3.97ATTNALWLKK21 pKa = 9.37WDD23 pKa = 3.76GVNVDD28 pKa = 3.43SMQKK32 pKa = 9.2QRR34 pKa = 11.84LWHH37 pKa = 6.53DD38 pKa = 3.63VQLVLSDD45 pKa = 3.63EE46 pKa = 4.47FEE48 pKa = 4.7GYY50 pKa = 10.67LRR52 pKa = 11.84VFVKK56 pKa = 10.06QEE58 pKa = 3.47PHH60 pKa = 6.12KK61 pKa = 9.95QRR63 pKa = 11.84KK64 pKa = 7.8MLDD67 pKa = 3.23KK68 pKa = 10.27RR69 pKa = 11.84WRR71 pKa = 11.84LIVASSLPVQVAWHH85 pKa = 5.3MLFYY89 pKa = 10.64QLNDD93 pKa = 3.6LEE95 pKa = 5.36IDD97 pKa = 3.44NSYY100 pKa = 10.43EE101 pKa = 3.84IPSQQGILLVKK112 pKa = 10.29GGWRR116 pKa = 11.84QYY118 pKa = 8.66YY119 pKa = 9.69NSWVEE124 pKa = 3.85RR125 pKa = 11.84GLVCGLDD132 pKa = 3.27KK133 pKa = 11.29SAWDD137 pKa = 3.15WTAPKK142 pKa = 9.41WAIDD146 pKa = 3.29MDD148 pKa = 4.09LEE150 pKa = 4.1LRR152 pKa = 11.84RR153 pKa = 11.84RR154 pKa = 11.84LGRR157 pKa = 11.84GDD159 pKa = 5.96KK160 pKa = 9.55MDD162 pKa = 3.23EE163 pKa = 3.47WHH165 pKa = 7.4RR166 pKa = 11.84IAKK169 pKa = 9.51SLYY172 pKa = 8.33RR173 pKa = 11.84RR174 pKa = 11.84MFEE177 pKa = 4.02DD178 pKa = 3.74PKK180 pKa = 11.14LILSDD185 pKa = 3.34GTVYY189 pKa = 9.3QQKK192 pKa = 9.44VPGIMKK198 pKa = 9.79SGCVNTISTNSHH210 pKa = 5.23CQVFVHH216 pKa = 6.51LAVCIEE222 pKa = 4.44NNLPMYY228 pKa = 8.36PLPVCCGDD236 pKa = 3.71DD237 pKa = 3.29TLQAVVHH244 pKa = 5.97VNDD247 pKa = 3.92LEE249 pKa = 4.07AYY251 pKa = 8.95RR252 pKa = 11.84RR253 pKa = 11.84FGVVVKK259 pKa = 10.47SASDD263 pKa = 3.48NIEE266 pKa = 4.12FVGHH270 pKa = 6.13EE271 pKa = 4.04FDD273 pKa = 5.04ARR275 pKa = 11.84GPQPMYY281 pKa = 10.39ILKK284 pKa = 9.89HH285 pKa = 5.31LKK287 pKa = 9.55KK288 pKa = 10.38VQYY291 pKa = 7.6TTDD294 pKa = 4.09EE295 pKa = 4.21ILSQYY300 pKa = 9.32LDD302 pKa = 3.25SMARR306 pKa = 11.84MYY308 pKa = 9.2THH310 pKa = 6.73SDD312 pKa = 4.06YY313 pKa = 11.45YY314 pKa = 10.5VVWEE318 pKa = 4.36TLAEE322 pKa = 4.01EE323 pKa = 4.46LGIDD327 pKa = 3.58LPMSRR332 pKa = 11.84EE333 pKa = 4.29SYY335 pKa = 9.19TYY337 pKa = 9.7WYY339 pKa = 10.23DD340 pKa = 3.65YY341 pKa = 11.57DD342 pKa = 3.38WW343 pKa = 5.71
MM1 pKa = 7.3TSSPGYY7 pKa = 9.38PYY9 pKa = 9.94MRR11 pKa = 11.84EE12 pKa = 3.97ATTNALWLKK21 pKa = 9.37WDD23 pKa = 3.76GVNVDD28 pKa = 3.43SMQKK32 pKa = 9.2QRR34 pKa = 11.84LWHH37 pKa = 6.53DD38 pKa = 3.63VQLVLSDD45 pKa = 3.63EE46 pKa = 4.47FEE48 pKa = 4.7GYY50 pKa = 10.67LRR52 pKa = 11.84VFVKK56 pKa = 10.06QEE58 pKa = 3.47PHH60 pKa = 6.12KK61 pKa = 9.95QRR63 pKa = 11.84KK64 pKa = 7.8MLDD67 pKa = 3.23KK68 pKa = 10.27RR69 pKa = 11.84WRR71 pKa = 11.84LIVASSLPVQVAWHH85 pKa = 5.3MLFYY89 pKa = 10.64QLNDD93 pKa = 3.6LEE95 pKa = 5.36IDD97 pKa = 3.44NSYY100 pKa = 10.43EE101 pKa = 3.84IPSQQGILLVKK112 pKa = 10.29GGWRR116 pKa = 11.84QYY118 pKa = 8.66YY119 pKa = 9.69NSWVEE124 pKa = 3.85RR125 pKa = 11.84GLVCGLDD132 pKa = 3.27KK133 pKa = 11.29SAWDD137 pKa = 3.15WTAPKK142 pKa = 9.41WAIDD146 pKa = 3.29MDD148 pKa = 4.09LEE150 pKa = 4.1LRR152 pKa = 11.84RR153 pKa = 11.84RR154 pKa = 11.84LGRR157 pKa = 11.84GDD159 pKa = 5.96KK160 pKa = 9.55MDD162 pKa = 3.23EE163 pKa = 3.47WHH165 pKa = 7.4RR166 pKa = 11.84IAKK169 pKa = 9.51SLYY172 pKa = 8.33RR173 pKa = 11.84RR174 pKa = 11.84MFEE177 pKa = 4.02DD178 pKa = 3.74PKK180 pKa = 11.14LILSDD185 pKa = 3.34GTVYY189 pKa = 9.3QQKK192 pKa = 9.44VPGIMKK198 pKa = 9.79SGCVNTISTNSHH210 pKa = 5.23CQVFVHH216 pKa = 6.51LAVCIEE222 pKa = 4.44NNLPMYY228 pKa = 8.36PLPVCCGDD236 pKa = 3.71DD237 pKa = 3.29TLQAVVHH244 pKa = 5.97VNDD247 pKa = 3.92LEE249 pKa = 4.07AYY251 pKa = 8.95RR252 pKa = 11.84RR253 pKa = 11.84FGVVVKK259 pKa = 10.47SASDD263 pKa = 3.48NIEE266 pKa = 4.12FVGHH270 pKa = 6.13EE271 pKa = 4.04FDD273 pKa = 5.04ARR275 pKa = 11.84GPQPMYY281 pKa = 10.39ILKK284 pKa = 9.89HH285 pKa = 5.31LKK287 pKa = 9.55KK288 pKa = 10.38VQYY291 pKa = 7.6TTDD294 pKa = 4.09EE295 pKa = 4.21ILSQYY300 pKa = 9.32LDD302 pKa = 3.25SMARR306 pKa = 11.84MYY308 pKa = 9.2THH310 pKa = 6.73SDD312 pKa = 4.06YY313 pKa = 11.45YY314 pKa = 10.5VVWEE318 pKa = 4.36TLAEE322 pKa = 4.01EE323 pKa = 4.46LGIDD327 pKa = 3.58LPMSRR332 pKa = 11.84EE333 pKa = 4.29SYY335 pKa = 9.19TYY337 pKa = 9.7WYY339 pKa = 10.23DD340 pKa = 3.65YY341 pKa = 11.57DD342 pKa = 3.38WW343 pKa = 5.71
Molecular weight: 40.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEH2|A0A1L3KEH2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 49 OX=1923237 PE=4 SV=1
MM1 pKa = 7.61ACFSDD6 pKa = 3.65MLIGFDD12 pKa = 3.47IEE14 pKa = 4.22PVEE17 pKa = 4.09RR18 pKa = 11.84RR19 pKa = 11.84SLSGVVMDD27 pKa = 4.54GTHH30 pKa = 6.46FVYY33 pKa = 10.65EE34 pKa = 4.23SAKK37 pKa = 11.03DD38 pKa = 3.62LVTTVAQTRR47 pKa = 11.84TKK49 pKa = 11.11GEE51 pKa = 4.0IVLGTMVFGGLVYY64 pKa = 10.35KK65 pKa = 10.39CYY67 pKa = 10.1EE68 pKa = 3.86YY69 pKa = 11.15SVPTKK74 pKa = 10.27ILRR77 pKa = 11.84CVSSVVPGYY86 pKa = 10.1RR87 pKa = 11.84WVKK90 pKa = 10.55ARR92 pKa = 11.84LGKK95 pKa = 10.56YY96 pKa = 9.69DD97 pKa = 3.66VVSAAPRR104 pKa = 11.84RR105 pKa = 11.84MDD107 pKa = 3.02SVLEE111 pKa = 3.95SRR113 pKa = 11.84RR114 pKa = 11.84TGSDD118 pKa = 3.58EE119 pKa = 4.31TPMTPPRR126 pKa = 11.84HH127 pKa = 4.96QAVVAIRR134 pKa = 11.84DD135 pKa = 3.62GDD137 pKa = 3.59IMRR140 pKa = 11.84VLGCAVRR147 pKa = 11.84FNNTLVGPDD156 pKa = 3.55HH157 pKa = 6.77VLGGTPDD164 pKa = 3.08VEE166 pKa = 4.53KK167 pKa = 10.4FVKK170 pKa = 10.75GSQSWVALASRR181 pKa = 11.84EE182 pKa = 4.17RR183 pKa = 11.84IPLDD187 pKa = 3.23TDD189 pKa = 3.93LVAIEE194 pKa = 4.68MSDD197 pKa = 3.73KK198 pKa = 11.05DD199 pKa = 3.71FASIGISACKK209 pKa = 8.43VAPMGNQTEE218 pKa = 4.63FAQIVGPDD226 pKa = 3.16AKK228 pKa = 10.44GTVGQLKK235 pKa = 10.14HH236 pKa = 6.82DD237 pKa = 5.54AICFGRR243 pKa = 11.84LVYY246 pKa = 10.34TGTTIQGYY254 pKa = 9.69SGAAYY259 pKa = 9.56TMGPKK264 pKa = 10.39VMGIHH269 pKa = 5.61QMGGAVNGGYY279 pKa = 9.45SAEE282 pKa = 4.24YY283 pKa = 10.13INNLLLVHH291 pKa = 6.61FKK293 pKa = 10.36QRR295 pKa = 11.84YY296 pKa = 5.82EE297 pKa = 4.39SSPEE301 pKa = 3.69WLLSQYY307 pKa = 10.38EE308 pKa = 4.24AGRR311 pKa = 11.84TMSWKK316 pKa = 9.19RR317 pKa = 11.84TGDD320 pKa = 3.26PSTYY324 pKa = 8.14QLKK327 pKa = 10.64VGGRR331 pKa = 11.84YY332 pKa = 9.86ALVDD336 pKa = 4.16RR337 pKa = 11.84DD338 pKa = 4.38DD339 pKa = 3.51MDD341 pKa = 4.14AAFGEE346 pKa = 4.38HH347 pKa = 5.65WRR349 pKa = 11.84DD350 pKa = 3.56SEE352 pKa = 4.63VIQNKK357 pKa = 9.26RR358 pKa = 11.84DD359 pKa = 3.44VRR361 pKa = 11.84HH362 pKa = 6.49RR363 pKa = 11.84DD364 pKa = 3.49YY365 pKa = 11.41EE366 pKa = 4.49STGAVANFLSNSAVSAPSSSGEE388 pKa = 4.01ANGSRR393 pKa = 11.84NLGALSVVDD402 pKa = 4.6DD403 pKa = 4.19SQGLEE408 pKa = 3.58MSGPQKK414 pKa = 11.03LMAEE418 pKa = 4.24YY419 pKa = 10.65AKK421 pKa = 10.77LSNAQRR427 pKa = 11.84KK428 pKa = 8.38RR429 pKa = 11.84FRR431 pKa = 11.84QLSNLSPRR439 pKa = 11.84PIEE442 pKa = 4.06ATAGQARR449 pKa = 11.84QTNSQSS455 pKa = 2.99
MM1 pKa = 7.61ACFSDD6 pKa = 3.65MLIGFDD12 pKa = 3.47IEE14 pKa = 4.22PVEE17 pKa = 4.09RR18 pKa = 11.84RR19 pKa = 11.84SLSGVVMDD27 pKa = 4.54GTHH30 pKa = 6.46FVYY33 pKa = 10.65EE34 pKa = 4.23SAKK37 pKa = 11.03DD38 pKa = 3.62LVTTVAQTRR47 pKa = 11.84TKK49 pKa = 11.11GEE51 pKa = 4.0IVLGTMVFGGLVYY64 pKa = 10.35KK65 pKa = 10.39CYY67 pKa = 10.1EE68 pKa = 3.86YY69 pKa = 11.15SVPTKK74 pKa = 10.27ILRR77 pKa = 11.84CVSSVVPGYY86 pKa = 10.1RR87 pKa = 11.84WVKK90 pKa = 10.55ARR92 pKa = 11.84LGKK95 pKa = 10.56YY96 pKa = 9.69DD97 pKa = 3.66VVSAAPRR104 pKa = 11.84RR105 pKa = 11.84MDD107 pKa = 3.02SVLEE111 pKa = 3.95SRR113 pKa = 11.84RR114 pKa = 11.84TGSDD118 pKa = 3.58EE119 pKa = 4.31TPMTPPRR126 pKa = 11.84HH127 pKa = 4.96QAVVAIRR134 pKa = 11.84DD135 pKa = 3.62GDD137 pKa = 3.59IMRR140 pKa = 11.84VLGCAVRR147 pKa = 11.84FNNTLVGPDD156 pKa = 3.55HH157 pKa = 6.77VLGGTPDD164 pKa = 3.08VEE166 pKa = 4.53KK167 pKa = 10.4FVKK170 pKa = 10.75GSQSWVALASRR181 pKa = 11.84EE182 pKa = 4.17RR183 pKa = 11.84IPLDD187 pKa = 3.23TDD189 pKa = 3.93LVAIEE194 pKa = 4.68MSDD197 pKa = 3.73KK198 pKa = 11.05DD199 pKa = 3.71FASIGISACKK209 pKa = 8.43VAPMGNQTEE218 pKa = 4.63FAQIVGPDD226 pKa = 3.16AKK228 pKa = 10.44GTVGQLKK235 pKa = 10.14HH236 pKa = 6.82DD237 pKa = 5.54AICFGRR243 pKa = 11.84LVYY246 pKa = 10.34TGTTIQGYY254 pKa = 9.69SGAAYY259 pKa = 9.56TMGPKK264 pKa = 10.39VMGIHH269 pKa = 5.61QMGGAVNGGYY279 pKa = 9.45SAEE282 pKa = 4.24YY283 pKa = 10.13INNLLLVHH291 pKa = 6.61FKK293 pKa = 10.36QRR295 pKa = 11.84YY296 pKa = 5.82EE297 pKa = 4.39SSPEE301 pKa = 3.69WLLSQYY307 pKa = 10.38EE308 pKa = 4.24AGRR311 pKa = 11.84TMSWKK316 pKa = 9.19RR317 pKa = 11.84TGDD320 pKa = 3.26PSTYY324 pKa = 8.14QLKK327 pKa = 10.64VGGRR331 pKa = 11.84YY332 pKa = 9.86ALVDD336 pKa = 4.16RR337 pKa = 11.84DD338 pKa = 4.38DD339 pKa = 3.51MDD341 pKa = 4.14AAFGEE346 pKa = 4.38HH347 pKa = 5.65WRR349 pKa = 11.84DD350 pKa = 3.56SEE352 pKa = 4.63VIQNKK357 pKa = 9.26RR358 pKa = 11.84DD359 pKa = 3.44VRR361 pKa = 11.84HH362 pKa = 6.49RR363 pKa = 11.84DD364 pKa = 3.49YY365 pKa = 11.41EE366 pKa = 4.49STGAVANFLSNSAVSAPSSSGEE388 pKa = 4.01ANGSRR393 pKa = 11.84NLGALSVVDD402 pKa = 4.6DD403 pKa = 4.19SQGLEE408 pKa = 3.58MSGPQKK414 pKa = 11.03LMAEE418 pKa = 4.24YY419 pKa = 10.65AKK421 pKa = 10.77LSNAQRR427 pKa = 11.84KK428 pKa = 8.38RR429 pKa = 11.84FRR431 pKa = 11.84QLSNLSPRR439 pKa = 11.84PIEE442 pKa = 4.06ATAGQARR449 pKa = 11.84QTNSQSS455 pKa = 2.99
Molecular weight: 49.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
798 |
343 |
455 |
399.0 |
45.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.642 ± 1.572 | 1.504 ± 0.17 |
6.642 ± 0.65 | 5.138 ± 0.278 |
2.632 ± 0.207 | 7.644 ± 1.661 |
2.256 ± 0.457 | 3.885 ± 0.136 |
4.762 ± 0.337 | 8.145 ± 1.225 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.759 ± 0.223 | 3.133 ± 0.051 |
4.135 ± 0.037 | 4.511 ± 0.308 |
6.516 ± 0.475 | 7.895 ± 1.026 |
4.762 ± 0.673 | 8.897 ± 0.307 |
2.381 ± 1.178 | 4.762 ± 0.943 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
