
Hubei sobemo-like virus 31
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEU2|A0A1L3KEU2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 31 OX=1923218 PE=4 SV=1
MM1 pKa = 7.42RR2 pKa = 11.84GEE4 pKa = 4.1RR5 pKa = 11.84AEE7 pKa = 4.17LTSLLYY13 pKa = 10.24HH14 pKa = 6.23AQLRR18 pKa = 11.84DD19 pKa = 3.85DD20 pKa = 4.05LVKK23 pKa = 10.88SITEE27 pKa = 3.95HH28 pKa = 6.03EE29 pKa = 4.29KK30 pKa = 11.21GILLNKK36 pKa = 10.33LEE38 pKa = 4.1MQFRR42 pKa = 11.84HH43 pKa = 6.06AKK45 pKa = 7.11WTIPSDD51 pKa = 3.51FMKK54 pKa = 10.29RR55 pKa = 11.84SHH57 pKa = 7.4FDD59 pKa = 3.11RR60 pKa = 11.84VLKK63 pKa = 10.75NIDD66 pKa = 3.43MTSSPGYY73 pKa = 9.63PYY75 pKa = 11.4LLVHH79 pKa = 6.5TNNASFFNCKK89 pKa = 10.0EE90 pKa = 3.72GVLDD94 pKa = 4.83VEE96 pKa = 5.13RR97 pKa = 11.84ADD99 pKa = 4.36QIWNRR104 pKa = 11.84VNSYY108 pKa = 10.43IEE110 pKa = 4.45NKK112 pKa = 10.4LSDD115 pKa = 4.82PIRR118 pKa = 11.84LFVKK122 pKa = 10.06PEE124 pKa = 3.61PHH126 pKa = 6.59NEE128 pKa = 3.35RR129 pKa = 11.84KK130 pKa = 8.74MRR132 pKa = 11.84SGRR135 pKa = 11.84YY136 pKa = 8.78RR137 pKa = 11.84LISSVSIIDD146 pKa = 3.78QIIDD150 pKa = 3.12AMLFGEE156 pKa = 4.48MNEE159 pKa = 3.96IMIHH163 pKa = 5.35NWPMLPTRR171 pKa = 11.84VGWSPYY177 pKa = 8.94YY178 pKa = 10.62GGWKK182 pKa = 9.11QMPYY186 pKa = 11.0AKK188 pKa = 9.21TQMAIDD194 pKa = 3.54KK195 pKa = 7.54TAWDD199 pKa = 3.66WSVQGWLPEE208 pKa = 3.81IVLEE212 pKa = 4.59LRR214 pKa = 11.84KK215 pKa = 9.82RR216 pKa = 11.84LCRR219 pKa = 11.84NLTSDD224 pKa = 4.0WEE226 pKa = 4.2RR227 pKa = 11.84LANWRR232 pKa = 11.84YY233 pKa = 9.44KK234 pKa = 10.98ALFGNPTLVTSGGLFLRR251 pKa = 11.84QKK253 pKa = 10.47QSGVMKK259 pKa = 9.96SGCYY263 pKa = 7.83NTIIDD268 pKa = 4.11NSIMQIVLHH277 pKa = 6.79DD278 pKa = 4.65LVCQTLNIPIGSIIAMGDD296 pKa = 3.33DD297 pKa = 3.77TLQDD301 pKa = 3.27ALFGEE306 pKa = 4.37EE307 pKa = 3.76RR308 pKa = 11.84EE309 pKa = 4.33RR310 pKa = 11.84YY311 pKa = 9.54LEE313 pKa = 4.07EE314 pKa = 3.79LSRR317 pKa = 11.84YY318 pKa = 10.15CNVKK322 pKa = 9.21VCNVSNEE329 pKa = 3.87FCGMHH334 pKa = 6.21FNIDD338 pKa = 3.32GRR340 pKa = 11.84VEE342 pKa = 3.56PVYY345 pKa = 10.23RR346 pKa = 11.84AKK348 pKa = 10.39HH349 pKa = 5.35AYY351 pKa = 8.94KK352 pKa = 9.96ILYY355 pKa = 8.61MDD357 pKa = 5.24DD358 pKa = 4.51SISDD362 pKa = 3.79SMADD366 pKa = 3.53SYY368 pKa = 11.84TLMYY372 pKa = 9.91YY373 pKa = 10.44RR374 pKa = 11.84SRR376 pKa = 11.84LKK378 pKa = 10.82DD379 pKa = 4.03LIDD382 pKa = 3.42SLFRR386 pKa = 11.84KK387 pKa = 8.5MGCEE391 pKa = 3.9VRR393 pKa = 11.84SDD395 pKa = 3.73EE396 pKa = 4.21YY397 pKa = 11.19RR398 pKa = 11.84DD399 pKa = 3.93AIFGG403 pKa = 3.75
MM1 pKa = 7.42RR2 pKa = 11.84GEE4 pKa = 4.1RR5 pKa = 11.84AEE7 pKa = 4.17LTSLLYY13 pKa = 10.24HH14 pKa = 6.23AQLRR18 pKa = 11.84DD19 pKa = 3.85DD20 pKa = 4.05LVKK23 pKa = 10.88SITEE27 pKa = 3.95HH28 pKa = 6.03EE29 pKa = 4.29KK30 pKa = 11.21GILLNKK36 pKa = 10.33LEE38 pKa = 4.1MQFRR42 pKa = 11.84HH43 pKa = 6.06AKK45 pKa = 7.11WTIPSDD51 pKa = 3.51FMKK54 pKa = 10.29RR55 pKa = 11.84SHH57 pKa = 7.4FDD59 pKa = 3.11RR60 pKa = 11.84VLKK63 pKa = 10.75NIDD66 pKa = 3.43MTSSPGYY73 pKa = 9.63PYY75 pKa = 11.4LLVHH79 pKa = 6.5TNNASFFNCKK89 pKa = 10.0EE90 pKa = 3.72GVLDD94 pKa = 4.83VEE96 pKa = 5.13RR97 pKa = 11.84ADD99 pKa = 4.36QIWNRR104 pKa = 11.84VNSYY108 pKa = 10.43IEE110 pKa = 4.45NKK112 pKa = 10.4LSDD115 pKa = 4.82PIRR118 pKa = 11.84LFVKK122 pKa = 10.06PEE124 pKa = 3.61PHH126 pKa = 6.59NEE128 pKa = 3.35RR129 pKa = 11.84KK130 pKa = 8.74MRR132 pKa = 11.84SGRR135 pKa = 11.84YY136 pKa = 8.78RR137 pKa = 11.84LISSVSIIDD146 pKa = 3.78QIIDD150 pKa = 3.12AMLFGEE156 pKa = 4.48MNEE159 pKa = 3.96IMIHH163 pKa = 5.35NWPMLPTRR171 pKa = 11.84VGWSPYY177 pKa = 8.94YY178 pKa = 10.62GGWKK182 pKa = 9.11QMPYY186 pKa = 11.0AKK188 pKa = 9.21TQMAIDD194 pKa = 3.54KK195 pKa = 7.54TAWDD199 pKa = 3.66WSVQGWLPEE208 pKa = 3.81IVLEE212 pKa = 4.59LRR214 pKa = 11.84KK215 pKa = 9.82RR216 pKa = 11.84LCRR219 pKa = 11.84NLTSDD224 pKa = 4.0WEE226 pKa = 4.2RR227 pKa = 11.84LANWRR232 pKa = 11.84YY233 pKa = 9.44KK234 pKa = 10.98ALFGNPTLVTSGGLFLRR251 pKa = 11.84QKK253 pKa = 10.47QSGVMKK259 pKa = 9.96SGCYY263 pKa = 7.83NTIIDD268 pKa = 4.11NSIMQIVLHH277 pKa = 6.79DD278 pKa = 4.65LVCQTLNIPIGSIIAMGDD296 pKa = 3.33DD297 pKa = 3.77TLQDD301 pKa = 3.27ALFGEE306 pKa = 4.37EE307 pKa = 3.76RR308 pKa = 11.84EE309 pKa = 4.33RR310 pKa = 11.84YY311 pKa = 9.54LEE313 pKa = 4.07EE314 pKa = 3.79LSRR317 pKa = 11.84YY318 pKa = 10.15CNVKK322 pKa = 9.21VCNVSNEE329 pKa = 3.87FCGMHH334 pKa = 6.21FNIDD338 pKa = 3.32GRR340 pKa = 11.84VEE342 pKa = 3.56PVYY345 pKa = 10.23RR346 pKa = 11.84AKK348 pKa = 10.39HH349 pKa = 5.35AYY351 pKa = 8.94KK352 pKa = 9.96ILYY355 pKa = 8.61MDD357 pKa = 5.24DD358 pKa = 4.51SISDD362 pKa = 3.79SMADD366 pKa = 3.53SYY368 pKa = 11.84TLMYY372 pKa = 9.91YY373 pKa = 10.44RR374 pKa = 11.84SRR376 pKa = 11.84LKK378 pKa = 10.82DD379 pKa = 4.03LIDD382 pKa = 3.42SLFRR386 pKa = 11.84KK387 pKa = 8.5MGCEE391 pKa = 3.9VRR393 pKa = 11.84SDD395 pKa = 3.73EE396 pKa = 4.21YY397 pKa = 11.19RR398 pKa = 11.84DD399 pKa = 3.93AIFGG403 pKa = 3.75
Molecular weight: 47.1 kDa
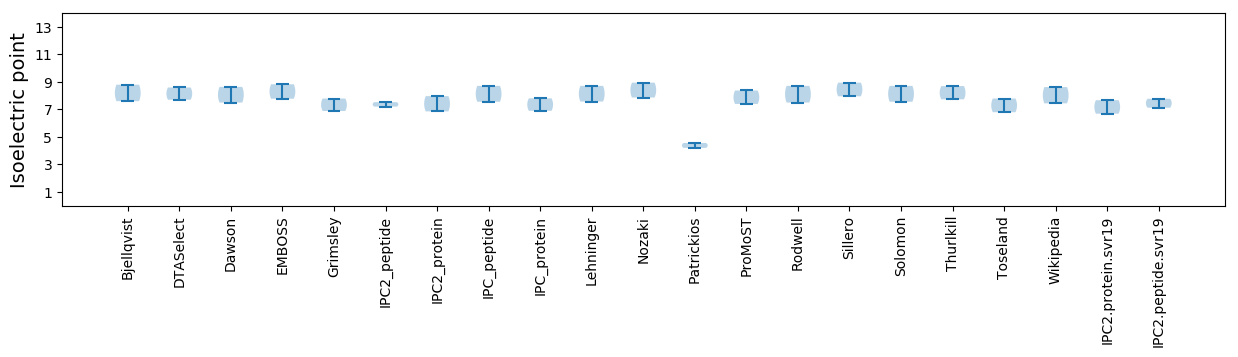
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEU2|A0A1L3KEU2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 31 OX=1923218 PE=4 SV=1
MM1 pKa = 7.3YY2 pKa = 10.64NVIDD6 pKa = 4.07AFTYY10 pKa = 10.68AVLGYY15 pKa = 10.44AATQAPGLSLAADD28 pKa = 4.11VIWRR32 pKa = 11.84SIKK35 pKa = 9.63WLFNVSFRR43 pKa = 11.84EE44 pKa = 3.93TKK46 pKa = 10.36AYY48 pKa = 9.87LKK50 pKa = 10.35PPTRR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 3.98VMRR59 pKa = 11.84DD60 pKa = 2.55WANRR64 pKa = 11.84LVEE67 pKa = 4.55RR68 pKa = 11.84GLAEE72 pKa = 3.86INAIQALEE80 pKa = 3.79WSIIGLVLVLLVLVWKK96 pKa = 10.32RR97 pKa = 11.84GRR99 pKa = 11.84RR100 pKa = 11.84TVHH103 pKa = 7.11RR104 pKa = 11.84IRR106 pKa = 11.84GIKK109 pKa = 9.07YY110 pKa = 7.75EE111 pKa = 3.96AMMANSHH118 pKa = 4.82FHH120 pKa = 5.99EE121 pKa = 5.3GKK123 pKa = 9.68PLEE126 pKa = 4.43CQVAILIPGLLTDD139 pKa = 3.66KK140 pKa = 11.1HH141 pKa = 6.39NGYY144 pKa = 9.83GFRR147 pKa = 11.84VEE149 pKa = 5.07DD150 pKa = 3.94FLVVPKK156 pKa = 10.57HH157 pKa = 4.9VVAGKK162 pKa = 10.43DD163 pKa = 3.34EE164 pKa = 4.31IMLKK168 pKa = 10.55NLFFPEE174 pKa = 4.13KK175 pKa = 10.43KK176 pKa = 10.74LMISTAGRR184 pKa = 11.84IDD186 pKa = 3.38SKK188 pKa = 11.69LNVDD192 pKa = 4.28LSYY195 pKa = 11.42ILLTKK200 pKa = 10.46KK201 pKa = 9.71SWSTVGVSKK210 pKa = 10.97SSIIAGTKK218 pKa = 10.35SLMQSVTIYY227 pKa = 10.44GPKK230 pKa = 10.01GVSEE234 pKa = 4.01GALRR238 pKa = 11.84HH239 pKa = 5.47TNMIGQLAYY248 pKa = 9.47TGSTIPGMSGSVYY261 pKa = 8.8TSEE264 pKa = 3.9GRR266 pKa = 11.84VLGMHH271 pKa = 6.66TGAFEE276 pKa = 4.03KK277 pKa = 10.5KK278 pKa = 10.05INVGLAGSMIKK289 pKa = 10.55EE290 pKa = 3.94EE291 pKa = 4.32VKK293 pKa = 10.9YY294 pKa = 9.36LTVVQLEE301 pKa = 4.29SSEE304 pKa = 4.22EE305 pKa = 3.99LLEE308 pKa = 4.23VAADD312 pKa = 3.53NGDD315 pKa = 3.6WNDD318 pKa = 3.17AMMNGYY324 pKa = 10.53VNDD327 pKa = 4.51KK328 pKa = 10.38YY329 pKa = 10.65SQRR332 pKa = 11.84QYY334 pKa = 11.61EE335 pKa = 3.98LDD337 pKa = 3.35QRR339 pKa = 11.84YY340 pKa = 9.57LEE342 pKa = 4.46DD343 pKa = 3.48KK344 pKa = 10.07KK345 pKa = 11.25AKK347 pKa = 10.08EE348 pKa = 4.35EE349 pKa = 4.02EE350 pKa = 3.94EE351 pKa = 3.82KK352 pKa = 10.79AYY354 pKa = 9.7TKK356 pKa = 10.54RR357 pKa = 11.84FYY359 pKa = 11.42ARR361 pKa = 11.84LASGEE366 pKa = 3.97IDD368 pKa = 4.05DD369 pKa = 4.15WTSDD373 pKa = 3.64DD374 pKa = 4.56FLGEE378 pKa = 4.15SAIKK382 pKa = 10.34LARR385 pKa = 11.84QLVEE389 pKa = 3.5RR390 pKa = 11.84ADD392 pKa = 4.1RR393 pKa = 11.84KK394 pKa = 10.65KK395 pKa = 9.95KK396 pKa = 8.48TIKK399 pKa = 10.66AEE401 pKa = 4.04SSDD404 pKa = 4.07GEE406 pKa = 4.72DD407 pKa = 4.66IPLQDD412 pKa = 3.02MALYY416 pKa = 10.33RR417 pKa = 11.84RR418 pKa = 11.84LTDD421 pKa = 3.14MEE423 pKa = 4.04ATIEE427 pKa = 3.99RR428 pKa = 11.84LAEE431 pKa = 3.66RR432 pKa = 11.84LAFVEE437 pKa = 4.16NRR439 pKa = 11.84LCRR442 pKa = 11.84EE443 pKa = 3.67TAMGQVVGYY452 pKa = 10.19RR453 pKa = 11.84CDD455 pKa = 3.3EE456 pKa = 4.26CRR458 pKa = 11.84TSCTTAEE465 pKa = 4.15KK466 pKa = 11.03LEE468 pKa = 4.04NHH470 pKa = 6.6KK471 pKa = 10.54KK472 pKa = 9.28AAHH475 pKa = 6.14PKK477 pKa = 8.74VEE479 pKa = 4.58KK480 pKa = 10.55YY481 pKa = 9.28PCPSCDD487 pKa = 3.42TVCNTQAKK495 pKa = 9.62LSNHH499 pKa = 5.75MLSNHH504 pKa = 5.51IPKK507 pKa = 9.72IEE509 pKa = 4.52LYY511 pKa = 10.16QCPLCNDD518 pKa = 3.58RR519 pKa = 11.84LEE521 pKa = 5.25KK522 pKa = 9.94EE523 pKa = 4.61ALMQHH528 pKa = 6.31ISKK531 pKa = 9.16HH532 pKa = 4.22QPEE535 pKa = 4.29AAEE538 pKa = 3.95EE539 pKa = 4.01AKK541 pKa = 10.67EE542 pKa = 3.8AVAFLGRR549 pKa = 11.84EE550 pKa = 3.69RR551 pKa = 11.84SSNQSSRR558 pKa = 11.84NGSTSWSAEE567 pKa = 3.75RR568 pKa = 11.84PRR570 pKa = 11.84QADD573 pKa = 3.08QPSPYY578 pKa = 10.08RR579 pKa = 11.84STTEE583 pKa = 3.62YY584 pKa = 10.82LKK586 pKa = 10.77KK587 pKa = 10.43KK588 pKa = 9.7KK589 pKa = 9.67PSSGKK594 pKa = 8.77PQQ596 pKa = 3.09
MM1 pKa = 7.3YY2 pKa = 10.64NVIDD6 pKa = 4.07AFTYY10 pKa = 10.68AVLGYY15 pKa = 10.44AATQAPGLSLAADD28 pKa = 4.11VIWRR32 pKa = 11.84SIKK35 pKa = 9.63WLFNVSFRR43 pKa = 11.84EE44 pKa = 3.93TKK46 pKa = 10.36AYY48 pKa = 9.87LKK50 pKa = 10.35PPTRR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 3.98VMRR59 pKa = 11.84DD60 pKa = 2.55WANRR64 pKa = 11.84LVEE67 pKa = 4.55RR68 pKa = 11.84GLAEE72 pKa = 3.86INAIQALEE80 pKa = 3.79WSIIGLVLVLLVLVWKK96 pKa = 10.32RR97 pKa = 11.84GRR99 pKa = 11.84RR100 pKa = 11.84TVHH103 pKa = 7.11RR104 pKa = 11.84IRR106 pKa = 11.84GIKK109 pKa = 9.07YY110 pKa = 7.75EE111 pKa = 3.96AMMANSHH118 pKa = 4.82FHH120 pKa = 5.99EE121 pKa = 5.3GKK123 pKa = 9.68PLEE126 pKa = 4.43CQVAILIPGLLTDD139 pKa = 3.66KK140 pKa = 11.1HH141 pKa = 6.39NGYY144 pKa = 9.83GFRR147 pKa = 11.84VEE149 pKa = 5.07DD150 pKa = 3.94FLVVPKK156 pKa = 10.57HH157 pKa = 4.9VVAGKK162 pKa = 10.43DD163 pKa = 3.34EE164 pKa = 4.31IMLKK168 pKa = 10.55NLFFPEE174 pKa = 4.13KK175 pKa = 10.43KK176 pKa = 10.74LMISTAGRR184 pKa = 11.84IDD186 pKa = 3.38SKK188 pKa = 11.69LNVDD192 pKa = 4.28LSYY195 pKa = 11.42ILLTKK200 pKa = 10.46KK201 pKa = 9.71SWSTVGVSKK210 pKa = 10.97SSIIAGTKK218 pKa = 10.35SLMQSVTIYY227 pKa = 10.44GPKK230 pKa = 10.01GVSEE234 pKa = 4.01GALRR238 pKa = 11.84HH239 pKa = 5.47TNMIGQLAYY248 pKa = 9.47TGSTIPGMSGSVYY261 pKa = 8.8TSEE264 pKa = 3.9GRR266 pKa = 11.84VLGMHH271 pKa = 6.66TGAFEE276 pKa = 4.03KK277 pKa = 10.5KK278 pKa = 10.05INVGLAGSMIKK289 pKa = 10.55EE290 pKa = 3.94EE291 pKa = 4.32VKK293 pKa = 10.9YY294 pKa = 9.36LTVVQLEE301 pKa = 4.29SSEE304 pKa = 4.22EE305 pKa = 3.99LLEE308 pKa = 4.23VAADD312 pKa = 3.53NGDD315 pKa = 3.6WNDD318 pKa = 3.17AMMNGYY324 pKa = 10.53VNDD327 pKa = 4.51KK328 pKa = 10.38YY329 pKa = 10.65SQRR332 pKa = 11.84QYY334 pKa = 11.61EE335 pKa = 3.98LDD337 pKa = 3.35QRR339 pKa = 11.84YY340 pKa = 9.57LEE342 pKa = 4.46DD343 pKa = 3.48KK344 pKa = 10.07KK345 pKa = 11.25AKK347 pKa = 10.08EE348 pKa = 4.35EE349 pKa = 4.02EE350 pKa = 3.94EE351 pKa = 3.82KK352 pKa = 10.79AYY354 pKa = 9.7TKK356 pKa = 10.54RR357 pKa = 11.84FYY359 pKa = 11.42ARR361 pKa = 11.84LASGEE366 pKa = 3.97IDD368 pKa = 4.05DD369 pKa = 4.15WTSDD373 pKa = 3.64DD374 pKa = 4.56FLGEE378 pKa = 4.15SAIKK382 pKa = 10.34LARR385 pKa = 11.84QLVEE389 pKa = 3.5RR390 pKa = 11.84ADD392 pKa = 4.1RR393 pKa = 11.84KK394 pKa = 10.65KK395 pKa = 9.95KK396 pKa = 8.48TIKK399 pKa = 10.66AEE401 pKa = 4.04SSDD404 pKa = 4.07GEE406 pKa = 4.72DD407 pKa = 4.66IPLQDD412 pKa = 3.02MALYY416 pKa = 10.33RR417 pKa = 11.84RR418 pKa = 11.84LTDD421 pKa = 3.14MEE423 pKa = 4.04ATIEE427 pKa = 3.99RR428 pKa = 11.84LAEE431 pKa = 3.66RR432 pKa = 11.84LAFVEE437 pKa = 4.16NRR439 pKa = 11.84LCRR442 pKa = 11.84EE443 pKa = 3.67TAMGQVVGYY452 pKa = 10.19RR453 pKa = 11.84CDD455 pKa = 3.3EE456 pKa = 4.26CRR458 pKa = 11.84TSCTTAEE465 pKa = 4.15KK466 pKa = 11.03LEE468 pKa = 4.04NHH470 pKa = 6.6KK471 pKa = 10.54KK472 pKa = 9.28AAHH475 pKa = 6.14PKK477 pKa = 8.74VEE479 pKa = 4.58KK480 pKa = 10.55YY481 pKa = 9.28PCPSCDD487 pKa = 3.42TVCNTQAKK495 pKa = 9.62LSNHH499 pKa = 5.75MLSNHH504 pKa = 5.51IPKK507 pKa = 9.72IEE509 pKa = 4.52LYY511 pKa = 10.16QCPLCNDD518 pKa = 3.58RR519 pKa = 11.84LEE521 pKa = 5.25KK522 pKa = 9.94EE523 pKa = 4.61ALMQHH528 pKa = 6.31ISKK531 pKa = 9.16HH532 pKa = 4.22QPEE535 pKa = 4.29AAEE538 pKa = 3.95EE539 pKa = 4.01AKK541 pKa = 10.67EE542 pKa = 3.8AVAFLGRR549 pKa = 11.84EE550 pKa = 3.69RR551 pKa = 11.84SSNQSSRR558 pKa = 11.84NGSTSWSAEE567 pKa = 3.75RR568 pKa = 11.84PRR570 pKa = 11.84QADD573 pKa = 3.08QPSPYY578 pKa = 10.08RR579 pKa = 11.84STTEE583 pKa = 3.62YY584 pKa = 10.82LKK586 pKa = 10.77KK587 pKa = 10.43KK588 pKa = 9.7KK589 pKa = 9.67PSSGKK594 pKa = 8.77PQQ596 pKa = 3.09
Molecular weight: 67.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
999 |
403 |
596 |
499.5 |
57.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.707 ± 1.39 | 1.802 ± 0.102 |
5.405 ± 0.723 | 7.407 ± 0.811 |
2.703 ± 0.431 | 5.806 ± 0.194 |
2.302 ± 0.1 | 5.906 ± 0.721 |
7.007 ± 0.865 | 9.409 ± 0.15 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.704 ± 0.565 | 4.304 ± 0.507 |
3.504 ± 0.017 | 3.203 ± 0.126 |
6.807 ± 0.356 | 7.508 ± 0.035 |
4.605 ± 0.493 | 5.806 ± 0.332 |
1.902 ± 0.324 | 4.204 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
