
Botrytis cinerea hypovirus 1 satellite-like RNA
Taxonomy: Viruses; Satellites; RNA satellites
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
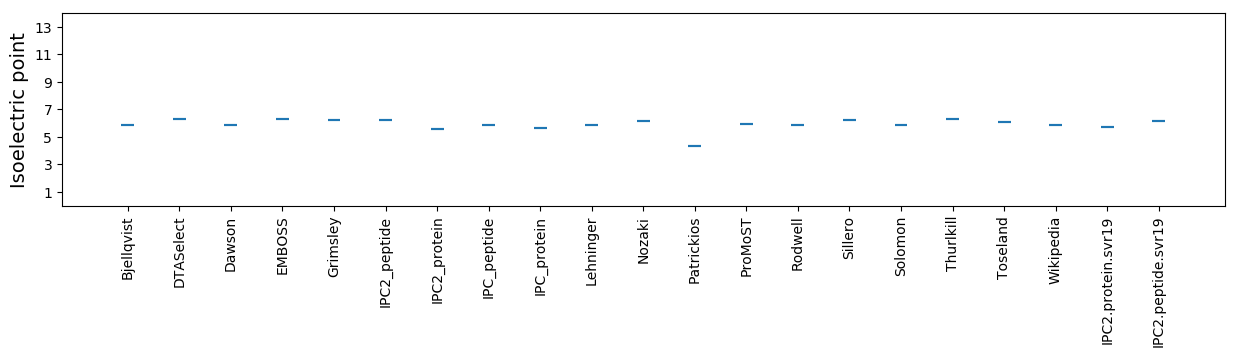
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U9NJB8|A0A2U9NJB8_9VIRU Uncharacterized protein OS=Botrytis cinerea hypovirus 1 satellite-like RNA OX=2219109 PE=4 SV=1
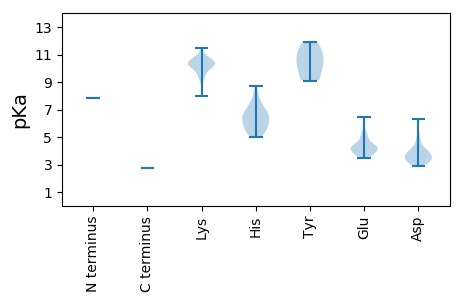
MM1 pKa = 7.81NIKK4 pKa = 10.16NKK6 pKa = 10.48NNFLDD11 pKa = 3.64NSSRR15 pKa = 11.84SLASTSSARR24 pKa = 11.84DD25 pKa = 3.33AKK27 pKa = 10.35PDD29 pKa = 3.67RR30 pKa = 11.84EE31 pKa = 4.23HH32 pKa = 7.52PEE34 pKa = 3.48QKK36 pKa = 10.37LRR38 pKa = 11.84LEE40 pKa = 4.3VQRR43 pKa = 11.84AEE45 pKa = 4.29KK46 pKa = 10.42PPVRR50 pKa = 11.84PSVFAGTEE58 pKa = 3.84HH59 pKa = 6.58QEE61 pKa = 3.91QDD63 pKa = 3.1GTQHH67 pKa = 5.59VRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.89HH72 pKa = 6.45HH73 pKa = 5.9GAHH76 pKa = 5.87SRR78 pKa = 11.84EE79 pKa = 3.89HH80 pKa = 6.59SKK82 pKa = 10.74EE83 pKa = 3.74SRR85 pKa = 11.84AGRR88 pKa = 11.84SSSHH92 pKa = 5.54SRR94 pKa = 11.84RR95 pKa = 11.84HH96 pKa = 5.96DD97 pKa = 3.86SEE99 pKa = 3.84HH100 pKa = 6.46HH101 pKa = 5.0RR102 pKa = 11.84TPRR105 pKa = 11.84ASSRR109 pKa = 11.84AEE111 pKa = 3.55SRR113 pKa = 11.84SRR115 pKa = 11.84IEE117 pKa = 4.41GKK119 pKa = 8.01TAADD123 pKa = 3.81KK124 pKa = 10.63GFCYY128 pKa = 10.07AALFLRR134 pKa = 11.84EE135 pKa = 4.24KK136 pKa = 10.02RR137 pKa = 11.84DD138 pKa = 3.43EE139 pKa = 4.4VIRR142 pKa = 11.84AIATDD147 pKa = 3.97YY148 pKa = 10.42PQLNKK153 pKa = 9.47VRR155 pKa = 11.84RR156 pKa = 11.84FADD159 pKa = 3.24THH161 pKa = 5.8KK162 pKa = 10.67AWVKK166 pKa = 10.46DD167 pKa = 3.04GHH169 pKa = 5.95ITVTAAIGFHH179 pKa = 6.67AGPNGEE185 pKa = 4.62LGNLKK190 pKa = 9.75PLSIHH195 pKa = 4.99VTSGGDD201 pKa = 3.27VSIKK205 pKa = 10.48EE206 pKa = 4.3FVASDD211 pKa = 3.46SYY213 pKa = 11.89GSVKK217 pKa = 10.18IGGYY221 pKa = 10.32ADD223 pKa = 4.09EE224 pKa = 5.4EE225 pKa = 4.25EE226 pKa = 5.26LDD228 pKa = 5.96DD229 pKa = 4.31EE230 pKa = 5.11GAWEE234 pKa = 4.94DD235 pKa = 5.18EE236 pKa = 4.18DD237 pKa = 6.29DD238 pKa = 3.83GWEE241 pKa = 4.26DD242 pKa = 4.6DD243 pKa = 3.97EE244 pKa = 4.44PTPVVKK250 pKa = 10.5SSKK253 pKa = 9.45TVVSSYY259 pKa = 10.66KK260 pKa = 10.05ISPTSGGNLNWPSSSFHH277 pKa = 7.89SNMGSSSRR285 pKa = 11.84STPVTPVAQSLDD297 pKa = 3.15WSHH300 pKa = 6.76YY301 pKa = 9.21GLLHH305 pKa = 5.34QVEE308 pKa = 4.84PRR310 pKa = 11.84PIINPPLPVKK320 pKa = 10.72SPEE323 pKa = 3.89RR324 pKa = 11.84PQNISNSLDD333 pKa = 3.73FYY335 pKa = 11.37HH336 pKa = 7.21EE337 pKa = 4.37SSDD340 pKa = 3.68DD341 pKa = 3.33KK342 pKa = 11.49CYY344 pKa = 11.16DD345 pKa = 4.15EE346 pKa = 6.47LPDD349 pKa = 4.92DD350 pKa = 4.05YY351 pKa = 11.55VFEE354 pKa = 4.84EE355 pKa = 5.22GEE357 pKa = 4.18DD358 pKa = 3.34WGTLTWPLPSTGVVAAAMDD377 pKa = 3.8ATLDD381 pKa = 3.9TEE383 pKa = 4.32PLSAEE388 pKa = 3.6ISSPLEE394 pKa = 3.73KK395 pKa = 10.15TISDD399 pKa = 3.63AVNAVIDD406 pKa = 4.36DD407 pKa = 4.28VVDD410 pKa = 3.61SADD413 pKa = 3.45PVFAALEE420 pKa = 4.12AATQFRR426 pKa = 11.84PVASRR431 pKa = 11.84DD432 pKa = 3.25ITSLVPKK439 pKa = 10.6AVGTCLSALASGNQPVVVARR459 pKa = 11.84SGSTGIVGFINGLIEE474 pKa = 4.12VYY476 pKa = 9.11KK477 pKa = 9.76TIIVACDD484 pKa = 3.27NFDD487 pKa = 2.92KK488 pKa = 11.1SMRR491 pKa = 11.84VHH493 pKa = 6.86PGWNVVTSAEE503 pKa = 4.34HH504 pKa = 5.48IVNGRR509 pKa = 11.84VNLGEE514 pKa = 3.96TRR516 pKa = 11.84DD517 pKa = 3.85LGSLPEE523 pKa = 3.95IVKK526 pKa = 10.46GSFIVVVVDD535 pKa = 3.68FDD537 pKa = 3.03NWTVKK542 pKa = 10.26KK543 pKa = 10.08QALFSAFDD551 pKa = 3.54GKK553 pKa = 10.42IIRR556 pKa = 11.84TSSSLKK562 pKa = 10.25LRR564 pKa = 11.84STGKK568 pKa = 9.95RR569 pKa = 11.84VLIRR573 pKa = 11.84IPRR576 pKa = 11.84EE577 pKa = 3.94DD578 pKa = 3.73KK579 pKa = 10.13SCQIVVTKK587 pKa = 10.37DD588 pKa = 3.22YY589 pKa = 11.15SCVVQAGRR597 pKa = 11.84NGVFIFATDD606 pKa = 3.45HH607 pKa = 6.58DD608 pKa = 4.89VSDD611 pKa = 3.27KK612 pKa = 10.8FYY614 pKa = 10.29WYY616 pKa = 9.57ILKK619 pKa = 9.31ATVFPSWVKK628 pKa = 9.14RR629 pKa = 11.84WGVVQHH635 pKa = 6.52WDD637 pKa = 3.19MSYY640 pKa = 9.17LTPRR644 pKa = 11.84MWNNCLCDD652 pKa = 3.35VHH654 pKa = 8.74LGMQEE659 pKa = 3.59PALEE663 pKa = 4.44VNCFHH668 pKa = 7.39SVV670 pKa = 2.75
MM1 pKa = 7.81NIKK4 pKa = 10.16NKK6 pKa = 10.48NNFLDD11 pKa = 3.64NSSRR15 pKa = 11.84SLASTSSARR24 pKa = 11.84DD25 pKa = 3.33AKK27 pKa = 10.35PDD29 pKa = 3.67RR30 pKa = 11.84EE31 pKa = 4.23HH32 pKa = 7.52PEE34 pKa = 3.48QKK36 pKa = 10.37LRR38 pKa = 11.84LEE40 pKa = 4.3VQRR43 pKa = 11.84AEE45 pKa = 4.29KK46 pKa = 10.42PPVRR50 pKa = 11.84PSVFAGTEE58 pKa = 3.84HH59 pKa = 6.58QEE61 pKa = 3.91QDD63 pKa = 3.1GTQHH67 pKa = 5.59VRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.89HH72 pKa = 6.45HH73 pKa = 5.9GAHH76 pKa = 5.87SRR78 pKa = 11.84EE79 pKa = 3.89HH80 pKa = 6.59SKK82 pKa = 10.74EE83 pKa = 3.74SRR85 pKa = 11.84AGRR88 pKa = 11.84SSSHH92 pKa = 5.54SRR94 pKa = 11.84RR95 pKa = 11.84HH96 pKa = 5.96DD97 pKa = 3.86SEE99 pKa = 3.84HH100 pKa = 6.46HH101 pKa = 5.0RR102 pKa = 11.84TPRR105 pKa = 11.84ASSRR109 pKa = 11.84AEE111 pKa = 3.55SRR113 pKa = 11.84SRR115 pKa = 11.84IEE117 pKa = 4.41GKK119 pKa = 8.01TAADD123 pKa = 3.81KK124 pKa = 10.63GFCYY128 pKa = 10.07AALFLRR134 pKa = 11.84EE135 pKa = 4.24KK136 pKa = 10.02RR137 pKa = 11.84DD138 pKa = 3.43EE139 pKa = 4.4VIRR142 pKa = 11.84AIATDD147 pKa = 3.97YY148 pKa = 10.42PQLNKK153 pKa = 9.47VRR155 pKa = 11.84RR156 pKa = 11.84FADD159 pKa = 3.24THH161 pKa = 5.8KK162 pKa = 10.67AWVKK166 pKa = 10.46DD167 pKa = 3.04GHH169 pKa = 5.95ITVTAAIGFHH179 pKa = 6.67AGPNGEE185 pKa = 4.62LGNLKK190 pKa = 9.75PLSIHH195 pKa = 4.99VTSGGDD201 pKa = 3.27VSIKK205 pKa = 10.48EE206 pKa = 4.3FVASDD211 pKa = 3.46SYY213 pKa = 11.89GSVKK217 pKa = 10.18IGGYY221 pKa = 10.32ADD223 pKa = 4.09EE224 pKa = 5.4EE225 pKa = 4.25EE226 pKa = 5.26LDD228 pKa = 5.96DD229 pKa = 4.31EE230 pKa = 5.11GAWEE234 pKa = 4.94DD235 pKa = 5.18EE236 pKa = 4.18DD237 pKa = 6.29DD238 pKa = 3.83GWEE241 pKa = 4.26DD242 pKa = 4.6DD243 pKa = 3.97EE244 pKa = 4.44PTPVVKK250 pKa = 10.5SSKK253 pKa = 9.45TVVSSYY259 pKa = 10.66KK260 pKa = 10.05ISPTSGGNLNWPSSSFHH277 pKa = 7.89SNMGSSSRR285 pKa = 11.84STPVTPVAQSLDD297 pKa = 3.15WSHH300 pKa = 6.76YY301 pKa = 9.21GLLHH305 pKa = 5.34QVEE308 pKa = 4.84PRR310 pKa = 11.84PIINPPLPVKK320 pKa = 10.72SPEE323 pKa = 3.89RR324 pKa = 11.84PQNISNSLDD333 pKa = 3.73FYY335 pKa = 11.37HH336 pKa = 7.21EE337 pKa = 4.37SSDD340 pKa = 3.68DD341 pKa = 3.33KK342 pKa = 11.49CYY344 pKa = 11.16DD345 pKa = 4.15EE346 pKa = 6.47LPDD349 pKa = 4.92DD350 pKa = 4.05YY351 pKa = 11.55VFEE354 pKa = 4.84EE355 pKa = 5.22GEE357 pKa = 4.18DD358 pKa = 3.34WGTLTWPLPSTGVVAAAMDD377 pKa = 3.8ATLDD381 pKa = 3.9TEE383 pKa = 4.32PLSAEE388 pKa = 3.6ISSPLEE394 pKa = 3.73KK395 pKa = 10.15TISDD399 pKa = 3.63AVNAVIDD406 pKa = 4.36DD407 pKa = 4.28VVDD410 pKa = 3.61SADD413 pKa = 3.45PVFAALEE420 pKa = 4.12AATQFRR426 pKa = 11.84PVASRR431 pKa = 11.84DD432 pKa = 3.25ITSLVPKK439 pKa = 10.6AVGTCLSALASGNQPVVVARR459 pKa = 11.84SGSTGIVGFINGLIEE474 pKa = 4.12VYY476 pKa = 9.11KK477 pKa = 9.76TIIVACDD484 pKa = 3.27NFDD487 pKa = 2.92KK488 pKa = 11.1SMRR491 pKa = 11.84VHH493 pKa = 6.86PGWNVVTSAEE503 pKa = 4.34HH504 pKa = 5.48IVNGRR509 pKa = 11.84VNLGEE514 pKa = 3.96TRR516 pKa = 11.84DD517 pKa = 3.85LGSLPEE523 pKa = 3.95IVKK526 pKa = 10.46GSFIVVVVDD535 pKa = 3.68FDD537 pKa = 3.03NWTVKK542 pKa = 10.26KK543 pKa = 10.08QALFSAFDD551 pKa = 3.54GKK553 pKa = 10.42IIRR556 pKa = 11.84TSSSLKK562 pKa = 10.25LRR564 pKa = 11.84STGKK568 pKa = 9.95RR569 pKa = 11.84VLIRR573 pKa = 11.84IPRR576 pKa = 11.84EE577 pKa = 3.94DD578 pKa = 3.73KK579 pKa = 10.13SCQIVVTKK587 pKa = 10.37DD588 pKa = 3.22YY589 pKa = 11.15SCVVQAGRR597 pKa = 11.84NGVFIFATDD606 pKa = 3.45HH607 pKa = 6.58DD608 pKa = 4.89VSDD611 pKa = 3.27KK612 pKa = 10.8FYY614 pKa = 10.29WYY616 pKa = 9.57ILKK619 pKa = 9.31ATVFPSWVKK628 pKa = 9.14RR629 pKa = 11.84WGVVQHH635 pKa = 6.52WDD637 pKa = 3.19MSYY640 pKa = 9.17LTPRR644 pKa = 11.84MWNNCLCDD652 pKa = 3.35VHH654 pKa = 8.74LGMQEE659 pKa = 3.59PALEE663 pKa = 4.44VNCFHH668 pKa = 7.39SVV670 pKa = 2.75
Molecular weight: 74.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U9NJB8|A0A2U9NJB8_9VIRU Uncharacterized protein OS=Botrytis cinerea hypovirus 1 satellite-like RNA OX=2219109 PE=4 SV=1
MM1 pKa = 7.81NIKK4 pKa = 10.16NKK6 pKa = 10.48NNFLDD11 pKa = 3.64NSSRR15 pKa = 11.84SLASTSSARR24 pKa = 11.84DD25 pKa = 3.33AKK27 pKa = 10.35PDD29 pKa = 3.67RR30 pKa = 11.84EE31 pKa = 4.23HH32 pKa = 7.52PEE34 pKa = 3.48QKK36 pKa = 10.37LRR38 pKa = 11.84LEE40 pKa = 4.3VQRR43 pKa = 11.84AEE45 pKa = 4.29KK46 pKa = 10.42PPVRR50 pKa = 11.84PSVFAGTEE58 pKa = 3.84HH59 pKa = 6.58QEE61 pKa = 3.91QDD63 pKa = 3.1GTQHH67 pKa = 5.59VRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.89HH72 pKa = 6.45HH73 pKa = 5.9GAHH76 pKa = 5.87SRR78 pKa = 11.84EE79 pKa = 3.89HH80 pKa = 6.59SKK82 pKa = 10.74EE83 pKa = 3.74SRR85 pKa = 11.84AGRR88 pKa = 11.84SSSHH92 pKa = 5.54SRR94 pKa = 11.84RR95 pKa = 11.84HH96 pKa = 5.96DD97 pKa = 3.86SEE99 pKa = 3.84HH100 pKa = 6.46HH101 pKa = 5.0RR102 pKa = 11.84TPRR105 pKa = 11.84ASSRR109 pKa = 11.84AEE111 pKa = 3.55SRR113 pKa = 11.84SRR115 pKa = 11.84IEE117 pKa = 4.41GKK119 pKa = 8.01TAADD123 pKa = 3.81KK124 pKa = 10.63GFCYY128 pKa = 10.07AALFLRR134 pKa = 11.84EE135 pKa = 4.24KK136 pKa = 10.02RR137 pKa = 11.84DD138 pKa = 3.43EE139 pKa = 4.4VIRR142 pKa = 11.84AIATDD147 pKa = 3.97YY148 pKa = 10.42PQLNKK153 pKa = 9.47VRR155 pKa = 11.84RR156 pKa = 11.84FADD159 pKa = 3.24THH161 pKa = 5.8KK162 pKa = 10.67AWVKK166 pKa = 10.46DD167 pKa = 3.04GHH169 pKa = 5.95ITVTAAIGFHH179 pKa = 6.67AGPNGEE185 pKa = 4.62LGNLKK190 pKa = 9.75PLSIHH195 pKa = 4.99VTSGGDD201 pKa = 3.27VSIKK205 pKa = 10.48EE206 pKa = 4.3FVASDD211 pKa = 3.46SYY213 pKa = 11.89GSVKK217 pKa = 10.18IGGYY221 pKa = 10.32ADD223 pKa = 4.09EE224 pKa = 5.4EE225 pKa = 4.25EE226 pKa = 5.26LDD228 pKa = 5.96DD229 pKa = 4.31EE230 pKa = 5.11GAWEE234 pKa = 4.94DD235 pKa = 5.18EE236 pKa = 4.18DD237 pKa = 6.29DD238 pKa = 3.83GWEE241 pKa = 4.26DD242 pKa = 4.6DD243 pKa = 3.97EE244 pKa = 4.44PTPVVKK250 pKa = 10.5SSKK253 pKa = 9.45TVVSSYY259 pKa = 10.66KK260 pKa = 10.05ISPTSGGNLNWPSSSFHH277 pKa = 7.89SNMGSSSRR285 pKa = 11.84STPVTPVAQSLDD297 pKa = 3.15WSHH300 pKa = 6.76YY301 pKa = 9.21GLLHH305 pKa = 5.34QVEE308 pKa = 4.84PRR310 pKa = 11.84PIINPPLPVKK320 pKa = 10.72SPEE323 pKa = 3.89RR324 pKa = 11.84PQNISNSLDD333 pKa = 3.73FYY335 pKa = 11.37HH336 pKa = 7.21EE337 pKa = 4.37SSDD340 pKa = 3.68DD341 pKa = 3.33KK342 pKa = 11.49CYY344 pKa = 11.16DD345 pKa = 4.15EE346 pKa = 6.47LPDD349 pKa = 4.92DD350 pKa = 4.05YY351 pKa = 11.55VFEE354 pKa = 4.84EE355 pKa = 5.22GEE357 pKa = 4.18DD358 pKa = 3.34WGTLTWPLPSTGVVAAAMDD377 pKa = 3.8ATLDD381 pKa = 3.9TEE383 pKa = 4.32PLSAEE388 pKa = 3.6ISSPLEE394 pKa = 3.73KK395 pKa = 10.15TISDD399 pKa = 3.63AVNAVIDD406 pKa = 4.36DD407 pKa = 4.28VVDD410 pKa = 3.61SADD413 pKa = 3.45PVFAALEE420 pKa = 4.12AATQFRR426 pKa = 11.84PVASRR431 pKa = 11.84DD432 pKa = 3.25ITSLVPKK439 pKa = 10.6AVGTCLSALASGNQPVVVARR459 pKa = 11.84SGSTGIVGFINGLIEE474 pKa = 4.12VYY476 pKa = 9.11KK477 pKa = 9.76TIIVACDD484 pKa = 3.27NFDD487 pKa = 2.92KK488 pKa = 11.1SMRR491 pKa = 11.84VHH493 pKa = 6.86PGWNVVTSAEE503 pKa = 4.34HH504 pKa = 5.48IVNGRR509 pKa = 11.84VNLGEE514 pKa = 3.96TRR516 pKa = 11.84DD517 pKa = 3.85LGSLPEE523 pKa = 3.95IVKK526 pKa = 10.46GSFIVVVVDD535 pKa = 3.68FDD537 pKa = 3.03NWTVKK542 pKa = 10.26KK543 pKa = 10.08QALFSAFDD551 pKa = 3.54GKK553 pKa = 10.42IIRR556 pKa = 11.84TSSSLKK562 pKa = 10.25LRR564 pKa = 11.84STGKK568 pKa = 9.95RR569 pKa = 11.84VLIRR573 pKa = 11.84IPRR576 pKa = 11.84EE577 pKa = 3.94DD578 pKa = 3.73KK579 pKa = 10.13SCQIVVTKK587 pKa = 10.37DD588 pKa = 3.22YY589 pKa = 11.15SCVVQAGRR597 pKa = 11.84NGVFIFATDD606 pKa = 3.45HH607 pKa = 6.58DD608 pKa = 4.89VSDD611 pKa = 3.27KK612 pKa = 10.8FYY614 pKa = 10.29WYY616 pKa = 9.57ILKK619 pKa = 9.31ATVFPSWVKK628 pKa = 9.14RR629 pKa = 11.84WGVVQHH635 pKa = 6.52WDD637 pKa = 3.19MSYY640 pKa = 9.17LTPRR644 pKa = 11.84MWNNCLCDD652 pKa = 3.35VHH654 pKa = 8.74LGMQEE659 pKa = 3.59PALEE663 pKa = 4.44VNCFHH668 pKa = 7.39SVV670 pKa = 2.75
MM1 pKa = 7.81NIKK4 pKa = 10.16NKK6 pKa = 10.48NNFLDD11 pKa = 3.64NSSRR15 pKa = 11.84SLASTSSARR24 pKa = 11.84DD25 pKa = 3.33AKK27 pKa = 10.35PDD29 pKa = 3.67RR30 pKa = 11.84EE31 pKa = 4.23HH32 pKa = 7.52PEE34 pKa = 3.48QKK36 pKa = 10.37LRR38 pKa = 11.84LEE40 pKa = 4.3VQRR43 pKa = 11.84AEE45 pKa = 4.29KK46 pKa = 10.42PPVRR50 pKa = 11.84PSVFAGTEE58 pKa = 3.84HH59 pKa = 6.58QEE61 pKa = 3.91QDD63 pKa = 3.1GTQHH67 pKa = 5.59VRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.89HH72 pKa = 6.45HH73 pKa = 5.9GAHH76 pKa = 5.87SRR78 pKa = 11.84EE79 pKa = 3.89HH80 pKa = 6.59SKK82 pKa = 10.74EE83 pKa = 3.74SRR85 pKa = 11.84AGRR88 pKa = 11.84SSSHH92 pKa = 5.54SRR94 pKa = 11.84RR95 pKa = 11.84HH96 pKa = 5.96DD97 pKa = 3.86SEE99 pKa = 3.84HH100 pKa = 6.46HH101 pKa = 5.0RR102 pKa = 11.84TPRR105 pKa = 11.84ASSRR109 pKa = 11.84AEE111 pKa = 3.55SRR113 pKa = 11.84SRR115 pKa = 11.84IEE117 pKa = 4.41GKK119 pKa = 8.01TAADD123 pKa = 3.81KK124 pKa = 10.63GFCYY128 pKa = 10.07AALFLRR134 pKa = 11.84EE135 pKa = 4.24KK136 pKa = 10.02RR137 pKa = 11.84DD138 pKa = 3.43EE139 pKa = 4.4VIRR142 pKa = 11.84AIATDD147 pKa = 3.97YY148 pKa = 10.42PQLNKK153 pKa = 9.47VRR155 pKa = 11.84RR156 pKa = 11.84FADD159 pKa = 3.24THH161 pKa = 5.8KK162 pKa = 10.67AWVKK166 pKa = 10.46DD167 pKa = 3.04GHH169 pKa = 5.95ITVTAAIGFHH179 pKa = 6.67AGPNGEE185 pKa = 4.62LGNLKK190 pKa = 9.75PLSIHH195 pKa = 4.99VTSGGDD201 pKa = 3.27VSIKK205 pKa = 10.48EE206 pKa = 4.3FVASDD211 pKa = 3.46SYY213 pKa = 11.89GSVKK217 pKa = 10.18IGGYY221 pKa = 10.32ADD223 pKa = 4.09EE224 pKa = 5.4EE225 pKa = 4.25EE226 pKa = 5.26LDD228 pKa = 5.96DD229 pKa = 4.31EE230 pKa = 5.11GAWEE234 pKa = 4.94DD235 pKa = 5.18EE236 pKa = 4.18DD237 pKa = 6.29DD238 pKa = 3.83GWEE241 pKa = 4.26DD242 pKa = 4.6DD243 pKa = 3.97EE244 pKa = 4.44PTPVVKK250 pKa = 10.5SSKK253 pKa = 9.45TVVSSYY259 pKa = 10.66KK260 pKa = 10.05ISPTSGGNLNWPSSSFHH277 pKa = 7.89SNMGSSSRR285 pKa = 11.84STPVTPVAQSLDD297 pKa = 3.15WSHH300 pKa = 6.76YY301 pKa = 9.21GLLHH305 pKa = 5.34QVEE308 pKa = 4.84PRR310 pKa = 11.84PIINPPLPVKK320 pKa = 10.72SPEE323 pKa = 3.89RR324 pKa = 11.84PQNISNSLDD333 pKa = 3.73FYY335 pKa = 11.37HH336 pKa = 7.21EE337 pKa = 4.37SSDD340 pKa = 3.68DD341 pKa = 3.33KK342 pKa = 11.49CYY344 pKa = 11.16DD345 pKa = 4.15EE346 pKa = 6.47LPDD349 pKa = 4.92DD350 pKa = 4.05YY351 pKa = 11.55VFEE354 pKa = 4.84EE355 pKa = 5.22GEE357 pKa = 4.18DD358 pKa = 3.34WGTLTWPLPSTGVVAAAMDD377 pKa = 3.8ATLDD381 pKa = 3.9TEE383 pKa = 4.32PLSAEE388 pKa = 3.6ISSPLEE394 pKa = 3.73KK395 pKa = 10.15TISDD399 pKa = 3.63AVNAVIDD406 pKa = 4.36DD407 pKa = 4.28VVDD410 pKa = 3.61SADD413 pKa = 3.45PVFAALEE420 pKa = 4.12AATQFRR426 pKa = 11.84PVASRR431 pKa = 11.84DD432 pKa = 3.25ITSLVPKK439 pKa = 10.6AVGTCLSALASGNQPVVVARR459 pKa = 11.84SGSTGIVGFINGLIEE474 pKa = 4.12VYY476 pKa = 9.11KK477 pKa = 9.76TIIVACDD484 pKa = 3.27NFDD487 pKa = 2.92KK488 pKa = 11.1SMRR491 pKa = 11.84VHH493 pKa = 6.86PGWNVVTSAEE503 pKa = 4.34HH504 pKa = 5.48IVNGRR509 pKa = 11.84VNLGEE514 pKa = 3.96TRR516 pKa = 11.84DD517 pKa = 3.85LGSLPEE523 pKa = 3.95IVKK526 pKa = 10.46GSFIVVVVDD535 pKa = 3.68FDD537 pKa = 3.03NWTVKK542 pKa = 10.26KK543 pKa = 10.08QALFSAFDD551 pKa = 3.54GKK553 pKa = 10.42IIRR556 pKa = 11.84TSSSLKK562 pKa = 10.25LRR564 pKa = 11.84STGKK568 pKa = 9.95RR569 pKa = 11.84VLIRR573 pKa = 11.84IPRR576 pKa = 11.84EE577 pKa = 3.94DD578 pKa = 3.73KK579 pKa = 10.13SCQIVVTKK587 pKa = 10.37DD588 pKa = 3.22YY589 pKa = 11.15SCVVQAGRR597 pKa = 11.84NGVFIFATDD606 pKa = 3.45HH607 pKa = 6.58DD608 pKa = 4.89VSDD611 pKa = 3.27KK612 pKa = 10.8FYY614 pKa = 10.29WYY616 pKa = 9.57ILKK619 pKa = 9.31ATVFPSWVKK628 pKa = 9.14RR629 pKa = 11.84WGVVQHH635 pKa = 6.52WDD637 pKa = 3.19MSYY640 pKa = 9.17LTPRR644 pKa = 11.84MWNNCLCDD652 pKa = 3.35VHH654 pKa = 8.74LGMQEE659 pKa = 3.59PALEE663 pKa = 4.44VNCFHH668 pKa = 7.39SVV670 pKa = 2.75
Molecular weight: 74.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
670 |
670 |
670 |
670.0 |
74.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.313 ± 0.0 | 1.343 ± 0.0 |
7.313 ± 0.0 | 6.269 ± 0.0 |
3.433 ± 0.0 | 6.269 ± 0.0 |
3.731 ± 0.0 | 4.776 ± 0.0 |
5.224 ± 0.0 | 5.97 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.045 ± 0.0 | 3.881 ± 0.0 |
5.522 ± 0.0 | 2.388 ± 0.0 |
5.97 ± 0.0 | 10.896 ± 0.0 |
5.224 ± 0.0 | 9.254 ± 0.0 |
2.09 ± 0.0 | 2.09 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
