
Human papillomavirus 197
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 24
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
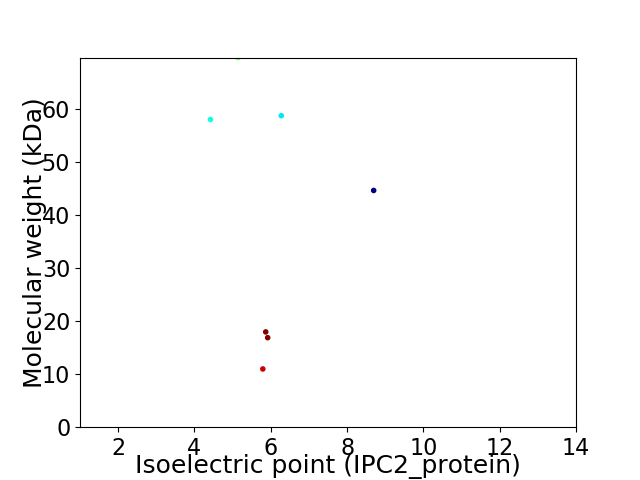
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A088FQG2|A0A088FQG2_9PAPI Replication protein E1 OS=Human papillomavirus 197 OX=1542134 GN=E1 PE=3 SV=1
MM1 pKa = 7.62EE2 pKa = 5.1PRR4 pKa = 11.84AKK6 pKa = 9.86RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.58RR10 pKa = 11.84DD11 pKa = 3.56TVDD14 pKa = 2.81NLYY17 pKa = 10.16RR18 pKa = 11.84QCKK21 pKa = 8.4LTGQCPDD28 pKa = 3.27DD29 pKa = 4.54VINKK33 pKa = 9.39VEE35 pKa = 4.16GTTLADD41 pKa = 3.77RR42 pKa = 11.84LLKK45 pKa = 10.5ILGSVIYY52 pKa = 10.6LGGLGIGTGRR62 pKa = 11.84GTGGATGYY70 pKa = 10.51RR71 pKa = 11.84PIGAQTPRR79 pKa = 11.84VTDD82 pKa = 3.45STPIRR87 pKa = 11.84PTIPVDD93 pKa = 3.35PLIPTDD99 pKa = 4.8IIPVDD104 pKa = 3.61PAGSSIVPLTDD115 pKa = 2.96AGIPEE120 pKa = 4.33DD121 pKa = 4.36VIIEE125 pKa = 4.3TGGSVVTAGPSDD137 pKa = 3.68PSVITTVDD145 pKa = 3.33PVSDD149 pKa = 3.69VTGVDD154 pKa = 3.68PQPTIITGEE163 pKa = 4.27DD164 pKa = 3.24DD165 pKa = 3.34TVAILDD171 pKa = 4.06VQPSTPAPKK180 pKa = 10.28KK181 pKa = 8.41VTTAVTQSASEE192 pKa = 4.02TLEE195 pKa = 3.73PSILLPIPEE204 pKa = 4.04SAANDD209 pKa = 2.96FHH211 pKa = 9.2IFVDD215 pKa = 4.2AQAIGEE221 pKa = 4.44TIGAEE226 pKa = 4.28TIPLEE231 pKa = 4.39DD232 pKa = 3.92FNEE235 pKa = 4.11IEE237 pKa = 4.2SFEE240 pKa = 4.43IEE242 pKa = 4.96DD243 pKa = 3.86IAPQTSTPEE252 pKa = 3.69QRR254 pKa = 11.84INRR257 pKa = 11.84AFQRR261 pKa = 11.84ARR263 pKa = 11.84SLYY266 pKa = 9.39NRR268 pKa = 11.84HH269 pKa = 6.11ISQIRR274 pKa = 11.84TRR276 pKa = 11.84NLDD279 pKa = 3.26FLGSVSRR286 pKa = 11.84AVQFEE291 pKa = 4.17FEE293 pKa = 4.25NPAFQGDD300 pKa = 3.53VTLQFEE306 pKa = 4.16QDD308 pKa = 3.48VRR310 pKa = 11.84EE311 pKa = 4.25VAAAPDD317 pKa = 3.4SDD319 pKa = 3.59FRR321 pKa = 11.84DD322 pKa = 3.05IRR324 pKa = 11.84TLSRR328 pKa = 11.84PQYY331 pKa = 9.19STTEE335 pKa = 3.9SGRR338 pKa = 11.84VRR340 pKa = 11.84VSRR343 pKa = 11.84LGRR346 pKa = 11.84RR347 pKa = 11.84GTILTRR353 pKa = 11.84SGTQIGEE360 pKa = 4.26NVHH363 pKa = 6.87FYY365 pKa = 11.42YY366 pKa = 10.74DD367 pKa = 3.97LSTIGSDD374 pKa = 2.9TADD377 pKa = 4.2AIEE380 pKa = 5.1LSTLGQQSGDD390 pKa = 3.44TTIVDD395 pKa = 4.26GLAEE399 pKa = 4.2SSFVDD404 pKa = 3.81VPNNTDD410 pKa = 2.74VAYY413 pKa = 10.43GEE415 pKa = 4.32EE416 pKa = 4.32TLFDD420 pKa = 4.08PQNEE424 pKa = 4.28EE425 pKa = 3.63FDD427 pKa = 3.96NLHH430 pKa = 6.65LVLTSTGRR438 pKa = 11.84RR439 pKa = 11.84GEE441 pKa = 3.91VDD443 pKa = 4.32NIPTLPPGTPLRR455 pKa = 11.84IFIDD459 pKa = 4.35DD460 pKa = 3.73YY461 pKa = 11.94GKK463 pKa = 10.52DD464 pKa = 3.7LIVNYY469 pKa = 9.63PMSYY473 pKa = 8.88DD474 pKa = 3.31TSTVIVPDD482 pKa = 3.98SAAHH486 pKa = 6.11PLEE489 pKa = 4.35PPILLDD495 pKa = 3.71YY496 pKa = 10.78LGSGYY501 pKa = 10.22YY502 pKa = 9.41LHH504 pKa = 7.36PSLWKK509 pKa = 8.62RR510 pKa = 11.84RR511 pKa = 11.84KK512 pKa = 9.38RR513 pKa = 11.84KK514 pKa = 10.03RR515 pKa = 11.84SDD517 pKa = 3.13TYY519 pKa = 11.64NSFTDD524 pKa = 5.12GIVDD528 pKa = 3.44ATEE531 pKa = 3.59WW532 pKa = 3.29
MM1 pKa = 7.62EE2 pKa = 5.1PRR4 pKa = 11.84AKK6 pKa = 9.86RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.58RR10 pKa = 11.84DD11 pKa = 3.56TVDD14 pKa = 2.81NLYY17 pKa = 10.16RR18 pKa = 11.84QCKK21 pKa = 8.4LTGQCPDD28 pKa = 3.27DD29 pKa = 4.54VINKK33 pKa = 9.39VEE35 pKa = 4.16GTTLADD41 pKa = 3.77RR42 pKa = 11.84LLKK45 pKa = 10.5ILGSVIYY52 pKa = 10.6LGGLGIGTGRR62 pKa = 11.84GTGGATGYY70 pKa = 10.51RR71 pKa = 11.84PIGAQTPRR79 pKa = 11.84VTDD82 pKa = 3.45STPIRR87 pKa = 11.84PTIPVDD93 pKa = 3.35PLIPTDD99 pKa = 4.8IIPVDD104 pKa = 3.61PAGSSIVPLTDD115 pKa = 2.96AGIPEE120 pKa = 4.33DD121 pKa = 4.36VIIEE125 pKa = 4.3TGGSVVTAGPSDD137 pKa = 3.68PSVITTVDD145 pKa = 3.33PVSDD149 pKa = 3.69VTGVDD154 pKa = 3.68PQPTIITGEE163 pKa = 4.27DD164 pKa = 3.24DD165 pKa = 3.34TVAILDD171 pKa = 4.06VQPSTPAPKK180 pKa = 10.28KK181 pKa = 8.41VTTAVTQSASEE192 pKa = 4.02TLEE195 pKa = 3.73PSILLPIPEE204 pKa = 4.04SAANDD209 pKa = 2.96FHH211 pKa = 9.2IFVDD215 pKa = 4.2AQAIGEE221 pKa = 4.44TIGAEE226 pKa = 4.28TIPLEE231 pKa = 4.39DD232 pKa = 3.92FNEE235 pKa = 4.11IEE237 pKa = 4.2SFEE240 pKa = 4.43IEE242 pKa = 4.96DD243 pKa = 3.86IAPQTSTPEE252 pKa = 3.69QRR254 pKa = 11.84INRR257 pKa = 11.84AFQRR261 pKa = 11.84ARR263 pKa = 11.84SLYY266 pKa = 9.39NRR268 pKa = 11.84HH269 pKa = 6.11ISQIRR274 pKa = 11.84TRR276 pKa = 11.84NLDD279 pKa = 3.26FLGSVSRR286 pKa = 11.84AVQFEE291 pKa = 4.17FEE293 pKa = 4.25NPAFQGDD300 pKa = 3.53VTLQFEE306 pKa = 4.16QDD308 pKa = 3.48VRR310 pKa = 11.84EE311 pKa = 4.25VAAAPDD317 pKa = 3.4SDD319 pKa = 3.59FRR321 pKa = 11.84DD322 pKa = 3.05IRR324 pKa = 11.84TLSRR328 pKa = 11.84PQYY331 pKa = 9.19STTEE335 pKa = 3.9SGRR338 pKa = 11.84VRR340 pKa = 11.84VSRR343 pKa = 11.84LGRR346 pKa = 11.84RR347 pKa = 11.84GTILTRR353 pKa = 11.84SGTQIGEE360 pKa = 4.26NVHH363 pKa = 6.87FYY365 pKa = 11.42YY366 pKa = 10.74DD367 pKa = 3.97LSTIGSDD374 pKa = 2.9TADD377 pKa = 4.2AIEE380 pKa = 5.1LSTLGQQSGDD390 pKa = 3.44TTIVDD395 pKa = 4.26GLAEE399 pKa = 4.2SSFVDD404 pKa = 3.81VPNNTDD410 pKa = 2.74VAYY413 pKa = 10.43GEE415 pKa = 4.32EE416 pKa = 4.32TLFDD420 pKa = 4.08PQNEE424 pKa = 4.28EE425 pKa = 3.63FDD427 pKa = 3.96NLHH430 pKa = 6.65LVLTSTGRR438 pKa = 11.84RR439 pKa = 11.84GEE441 pKa = 3.91VDD443 pKa = 4.32NIPTLPPGTPLRR455 pKa = 11.84IFIDD459 pKa = 4.35DD460 pKa = 3.73YY461 pKa = 11.94GKK463 pKa = 10.52DD464 pKa = 3.7LIVNYY469 pKa = 9.63PMSYY473 pKa = 8.88DD474 pKa = 3.31TSTVIVPDD482 pKa = 3.98SAAHH486 pKa = 6.11PLEE489 pKa = 4.35PPILLDD495 pKa = 3.71YY496 pKa = 10.78LGSGYY501 pKa = 10.22YY502 pKa = 9.41LHH504 pKa = 7.36PSLWKK509 pKa = 8.62RR510 pKa = 11.84RR511 pKa = 11.84KK512 pKa = 9.38RR513 pKa = 11.84KK514 pKa = 10.03RR515 pKa = 11.84SDD517 pKa = 3.13TYY519 pKa = 11.64NSFTDD524 pKa = 5.12GIVDD528 pKa = 3.44ATEE531 pKa = 3.59WW532 pKa = 3.29
Molecular weight: 57.93 kDa
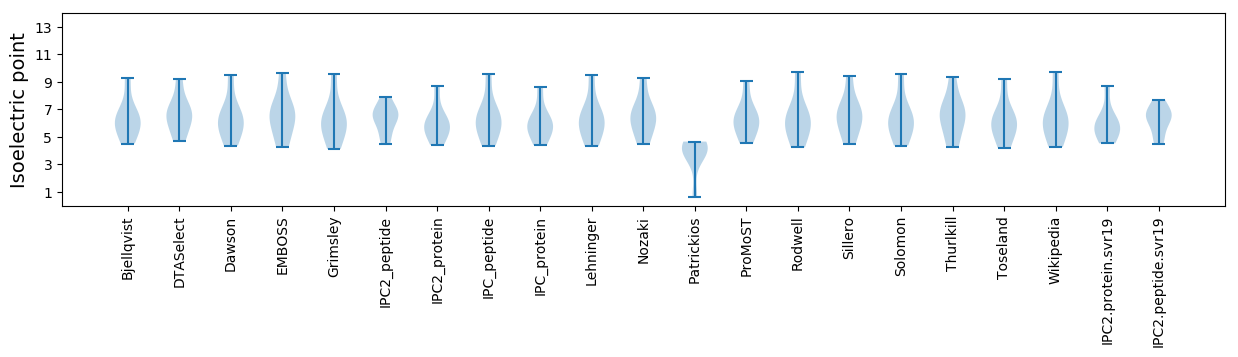
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A088FLI4|A0A088FLI4_9PAPI Protein E7 OS=Human papillomavirus 197 OX=1542134 GN=E7 PE=3 SV=1
MM1 pKa = 7.35EE2 pKa = 4.46WLAEE6 pKa = 4.05RR7 pKa = 11.84FNAVQEE13 pKa = 4.29TLLNLIEE20 pKa = 4.74QGAEE24 pKa = 3.98DD25 pKa = 5.21LDD27 pKa = 4.15SQITYY32 pKa = 9.18WNTVRR37 pKa = 11.84KK38 pKa = 9.28EE39 pKa = 4.05NVYY42 pKa = 9.6MYY44 pKa = 10.0YY45 pKa = 10.76AKK47 pKa = 10.38KK48 pKa = 10.51EE49 pKa = 4.11KK50 pKa = 9.33LTHH53 pKa = 6.89LGLQPLPVTAVSEE66 pKa = 4.49YY67 pKa = 10.03KK68 pKa = 10.48AKK70 pKa = 10.17QAIHH74 pKa = 6.46IVLLLEE80 pKa = 4.3SLKK83 pKa = 10.74KK84 pKa = 10.87SPFANEE90 pKa = 3.13AWTLQNTSAEE100 pKa = 4.34LLNTLPKK107 pKa = 10.77DD108 pKa = 3.88CFKK111 pKa = 10.93KK112 pKa = 10.62DD113 pKa = 3.42PYY115 pKa = 9.43TVHH118 pKa = 6.05VWFDD122 pKa = 3.5DD123 pKa = 3.44DD124 pKa = 4.76RR125 pKa = 11.84NNSFPYY131 pKa = 10.32VNWDD135 pKa = 3.58AIYY138 pKa = 10.77YY139 pKa = 10.08QDD141 pKa = 5.55AQGKK145 pKa = 4.24WHH147 pKa = 6.82KK148 pKa = 11.01VPGLVDD154 pKa = 3.71YY155 pKa = 11.46NGLYY159 pKa = 9.67YY160 pKa = 10.94NEE162 pKa = 4.3VGGDD166 pKa = 2.98RR167 pKa = 11.84VYY169 pKa = 10.74FALFDD174 pKa = 4.72ADD176 pKa = 3.5AQNYY180 pKa = 7.67GHH182 pKa = 6.65TGMWSVHH189 pKa = 5.54FKK191 pKa = 11.1HH192 pKa = 5.86EE193 pKa = 4.29TLSPISSSKK202 pKa = 10.62QSTSYY207 pKa = 10.5SGKK210 pKa = 8.0TSGVSTTTKK219 pKa = 10.32VPVPTAEE226 pKa = 4.07SPRR229 pKa = 11.84RR230 pKa = 11.84LQKK233 pKa = 10.74PEE235 pKa = 3.96VGSSTGEE242 pKa = 3.5KK243 pKa = 8.16TAIRR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84QQRR254 pKa = 11.84EE255 pKa = 4.02STPEE259 pKa = 4.0GEE261 pKa = 4.48PTTSSKK267 pKa = 10.27RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84GGGGGADD277 pKa = 3.24RR278 pKa = 11.84LGVSPEE284 pKa = 4.05AVGSRR289 pKa = 11.84HH290 pKa = 5.89RR291 pKa = 11.84SVTASHH297 pKa = 7.33LSRR300 pKa = 11.84LEE302 pKa = 3.74LLKK305 pKa = 11.06EE306 pKa = 4.12EE307 pKa = 5.08ARR309 pKa = 11.84DD310 pKa = 3.56PPIIIITGPANTLKK324 pKa = 10.22CWRR327 pKa = 11.84YY328 pKa = 9.66RR329 pKa = 11.84KK330 pKa = 9.84RR331 pKa = 11.84NSNASWFLGISTIFTWVGGCSTNEE355 pKa = 3.75QARR358 pKa = 11.84MLVAFKK364 pKa = 10.77NDD366 pKa = 3.27QQRR369 pKa = 11.84EE370 pKa = 4.09QFVKK374 pKa = 10.67YY375 pKa = 10.07IQLPKK380 pKa = 10.26QATFAYY386 pKa = 9.37GQLDD390 pKa = 3.49RR391 pKa = 11.84LL392 pKa = 4.02
MM1 pKa = 7.35EE2 pKa = 4.46WLAEE6 pKa = 4.05RR7 pKa = 11.84FNAVQEE13 pKa = 4.29TLLNLIEE20 pKa = 4.74QGAEE24 pKa = 3.98DD25 pKa = 5.21LDD27 pKa = 4.15SQITYY32 pKa = 9.18WNTVRR37 pKa = 11.84KK38 pKa = 9.28EE39 pKa = 4.05NVYY42 pKa = 9.6MYY44 pKa = 10.0YY45 pKa = 10.76AKK47 pKa = 10.38KK48 pKa = 10.51EE49 pKa = 4.11KK50 pKa = 9.33LTHH53 pKa = 6.89LGLQPLPVTAVSEE66 pKa = 4.49YY67 pKa = 10.03KK68 pKa = 10.48AKK70 pKa = 10.17QAIHH74 pKa = 6.46IVLLLEE80 pKa = 4.3SLKK83 pKa = 10.74KK84 pKa = 10.87SPFANEE90 pKa = 3.13AWTLQNTSAEE100 pKa = 4.34LLNTLPKK107 pKa = 10.77DD108 pKa = 3.88CFKK111 pKa = 10.93KK112 pKa = 10.62DD113 pKa = 3.42PYY115 pKa = 9.43TVHH118 pKa = 6.05VWFDD122 pKa = 3.5DD123 pKa = 3.44DD124 pKa = 4.76RR125 pKa = 11.84NNSFPYY131 pKa = 10.32VNWDD135 pKa = 3.58AIYY138 pKa = 10.77YY139 pKa = 10.08QDD141 pKa = 5.55AQGKK145 pKa = 4.24WHH147 pKa = 6.82KK148 pKa = 11.01VPGLVDD154 pKa = 3.71YY155 pKa = 11.46NGLYY159 pKa = 9.67YY160 pKa = 10.94NEE162 pKa = 4.3VGGDD166 pKa = 2.98RR167 pKa = 11.84VYY169 pKa = 10.74FALFDD174 pKa = 4.72ADD176 pKa = 3.5AQNYY180 pKa = 7.67GHH182 pKa = 6.65TGMWSVHH189 pKa = 5.54FKK191 pKa = 11.1HH192 pKa = 5.86EE193 pKa = 4.29TLSPISSSKK202 pKa = 10.62QSTSYY207 pKa = 10.5SGKK210 pKa = 8.0TSGVSTTTKK219 pKa = 10.32VPVPTAEE226 pKa = 4.07SPRR229 pKa = 11.84RR230 pKa = 11.84LQKK233 pKa = 10.74PEE235 pKa = 3.96VGSSTGEE242 pKa = 3.5KK243 pKa = 8.16TAIRR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84QQRR254 pKa = 11.84EE255 pKa = 4.02STPEE259 pKa = 4.0GEE261 pKa = 4.48PTTSSKK267 pKa = 10.27RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84GGGGGADD277 pKa = 3.24RR278 pKa = 11.84LGVSPEE284 pKa = 4.05AVGSRR289 pKa = 11.84HH290 pKa = 5.89RR291 pKa = 11.84SVTASHH297 pKa = 7.33LSRR300 pKa = 11.84LEE302 pKa = 3.74LLKK305 pKa = 11.06EE306 pKa = 4.12EE307 pKa = 5.08ARR309 pKa = 11.84DD310 pKa = 3.56PPIIIITGPANTLKK324 pKa = 10.22CWRR327 pKa = 11.84YY328 pKa = 9.66RR329 pKa = 11.84KK330 pKa = 9.84RR331 pKa = 11.84NSNASWFLGISTIFTWVGGCSTNEE355 pKa = 3.75QARR358 pKa = 11.84MLVAFKK364 pKa = 10.77NDD366 pKa = 3.27QQRR369 pKa = 11.84EE370 pKa = 4.09QFVKK374 pKa = 10.67YY375 pKa = 10.07IQLPKK380 pKa = 10.26QATFAYY386 pKa = 9.37GQLDD390 pKa = 3.49RR391 pKa = 11.84LL392 pKa = 4.02
Molecular weight: 44.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2445 |
94 |
607 |
349.3 |
39.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.849 ± 0.407 | 2.331 ± 0.766 |
6.83 ± 0.627 | 5.93 ± 0.369 |
4.294 ± 0.458 | 5.726 ± 0.736 |
1.84 ± 0.26 | 5.685 ± 0.627 |
5.235 ± 0.896 | 8.957 ± 0.581 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.513 ± 0.378 | 5.276 ± 0.703 |
5.726 ± 1.001 | 4.99 ± 0.36 |
5.726 ± 0.649 | 6.585 ± 0.395 |
6.462 ± 0.997 | 6.135 ± 0.395 |
1.309 ± 0.288 | 3.599 ± 0.369 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
