
Giant panda circovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 8.0
Get precalculated fractions of proteins
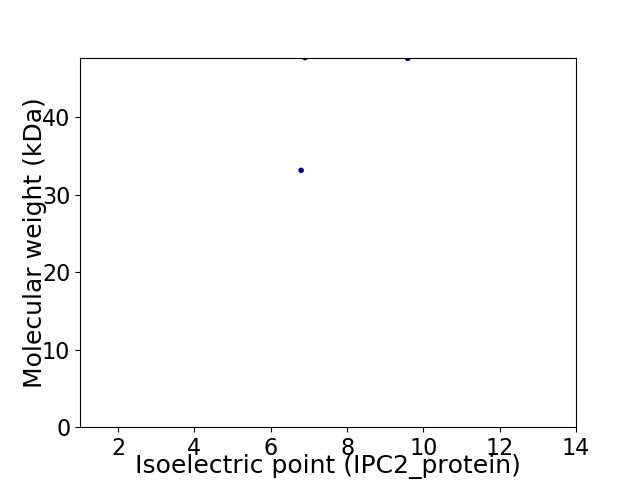
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A220IGS1|A0A220IGS1_9CIRC Capsid protein OS=Giant panda circovirus 1 OX=2016456 PE=4 SV=1
MM1 pKa = 7.16SQPEE5 pKa = 4.17PLLSRR10 pKa = 11.84SEE12 pKa = 4.01ASEE15 pKa = 4.31DD16 pKa = 3.37GSRR19 pKa = 11.84SSKK22 pKa = 9.74SRR24 pKa = 11.84RR25 pKa = 11.84QGRR28 pKa = 11.84YY29 pKa = 7.69WILTISCDD37 pKa = 3.89SNPWTPTALLPGQSYY52 pKa = 9.2VVGQQEE58 pKa = 4.29VGVGGFKK65 pKa = 9.95HH66 pKa = 4.96WQVMVAYY73 pKa = 8.97SKK75 pKa = 10.95KK76 pKa = 9.81VGLRR80 pKa = 11.84CVQRR84 pKa = 11.84DD85 pKa = 3.84FGSCHH90 pKa = 7.2AEE92 pKa = 3.75LTYY95 pKa = 10.89SDD97 pKa = 4.26SARR100 pKa = 11.84EE101 pKa = 3.99YY102 pKa = 10.6CRR104 pKa = 11.84KK105 pKa = 9.86EE106 pKa = 3.75DD107 pKa = 3.58TRR109 pKa = 11.84VEE111 pKa = 4.14GTQFEE116 pKa = 4.92FGTLATRR123 pKa = 11.84RR124 pKa = 11.84NSEE127 pKa = 4.0TDD129 pKa = 2.44WDD131 pKa = 4.46VIRR134 pKa = 11.84DD135 pKa = 3.75AAQLSDD141 pKa = 3.81LSGVPSDD148 pKa = 3.99IYY150 pKa = 10.6VRR152 pKa = 11.84CYY154 pKa = 10.06NQLRR158 pKa = 11.84RR159 pKa = 11.84IGQDD163 pKa = 2.71HH164 pKa = 6.92LRR166 pKa = 11.84PVGMEE171 pKa = 4.04RR172 pKa = 11.84TCHH175 pKa = 5.14VFWGRR180 pKa = 11.84TGTGKK185 pKa = 9.99SRR187 pKa = 11.84RR188 pKa = 11.84AWEE191 pKa = 3.97EE192 pKa = 3.63AGLCAYY198 pKa = 9.5PKK200 pKa = 10.71DD201 pKa = 4.16PNTKK205 pKa = 9.51FWDD208 pKa = 4.49GYY210 pKa = 9.96RR211 pKa = 11.84NHH213 pKa = 5.95EE214 pKa = 4.2HH215 pKa = 6.16VVIDD219 pKa = 3.79EE220 pKa = 3.96FRR222 pKa = 11.84GIINISNLLRR232 pKa = 11.84WLDD235 pKa = 3.8RR236 pKa = 11.84YY237 pKa = 9.87PVIVEE242 pKa = 3.99VKK244 pKa = 8.23GSSVVFCARR253 pKa = 11.84TIWITSNVNPEE264 pKa = 3.54QWYY267 pKa = 9.59PDD269 pKa = 3.59ADD271 pKa = 3.92QEE273 pKa = 4.72TKK275 pKa = 10.77DD276 pKa = 3.36ALLRR280 pKa = 11.84RR281 pKa = 11.84LTVVYY286 pKa = 9.55FEE288 pKa = 4.61
MM1 pKa = 7.16SQPEE5 pKa = 4.17PLLSRR10 pKa = 11.84SEE12 pKa = 4.01ASEE15 pKa = 4.31DD16 pKa = 3.37GSRR19 pKa = 11.84SSKK22 pKa = 9.74SRR24 pKa = 11.84RR25 pKa = 11.84QGRR28 pKa = 11.84YY29 pKa = 7.69WILTISCDD37 pKa = 3.89SNPWTPTALLPGQSYY52 pKa = 9.2VVGQQEE58 pKa = 4.29VGVGGFKK65 pKa = 9.95HH66 pKa = 4.96WQVMVAYY73 pKa = 8.97SKK75 pKa = 10.95KK76 pKa = 9.81VGLRR80 pKa = 11.84CVQRR84 pKa = 11.84DD85 pKa = 3.84FGSCHH90 pKa = 7.2AEE92 pKa = 3.75LTYY95 pKa = 10.89SDD97 pKa = 4.26SARR100 pKa = 11.84EE101 pKa = 3.99YY102 pKa = 10.6CRR104 pKa = 11.84KK105 pKa = 9.86EE106 pKa = 3.75DD107 pKa = 3.58TRR109 pKa = 11.84VEE111 pKa = 4.14GTQFEE116 pKa = 4.92FGTLATRR123 pKa = 11.84RR124 pKa = 11.84NSEE127 pKa = 4.0TDD129 pKa = 2.44WDD131 pKa = 4.46VIRR134 pKa = 11.84DD135 pKa = 3.75AAQLSDD141 pKa = 3.81LSGVPSDD148 pKa = 3.99IYY150 pKa = 10.6VRR152 pKa = 11.84CYY154 pKa = 10.06NQLRR158 pKa = 11.84RR159 pKa = 11.84IGQDD163 pKa = 2.71HH164 pKa = 6.92LRR166 pKa = 11.84PVGMEE171 pKa = 4.04RR172 pKa = 11.84TCHH175 pKa = 5.14VFWGRR180 pKa = 11.84TGTGKK185 pKa = 9.99SRR187 pKa = 11.84RR188 pKa = 11.84AWEE191 pKa = 3.97EE192 pKa = 3.63AGLCAYY198 pKa = 9.5PKK200 pKa = 10.71DD201 pKa = 4.16PNTKK205 pKa = 9.51FWDD208 pKa = 4.49GYY210 pKa = 9.96RR211 pKa = 11.84NHH213 pKa = 5.95EE214 pKa = 4.2HH215 pKa = 6.16VVIDD219 pKa = 3.79EE220 pKa = 3.96FRR222 pKa = 11.84GIINISNLLRR232 pKa = 11.84WLDD235 pKa = 3.8RR236 pKa = 11.84YY237 pKa = 9.87PVIVEE242 pKa = 3.99VKK244 pKa = 8.23GSSVVFCARR253 pKa = 11.84TIWITSNVNPEE264 pKa = 3.54QWYY267 pKa = 9.59PDD269 pKa = 3.59ADD271 pKa = 3.92QEE273 pKa = 4.72TKK275 pKa = 10.77DD276 pKa = 3.36ALLRR280 pKa = 11.84RR281 pKa = 11.84LTVVYY286 pKa = 9.55FEE288 pKa = 4.61
Molecular weight: 33.13 kDa
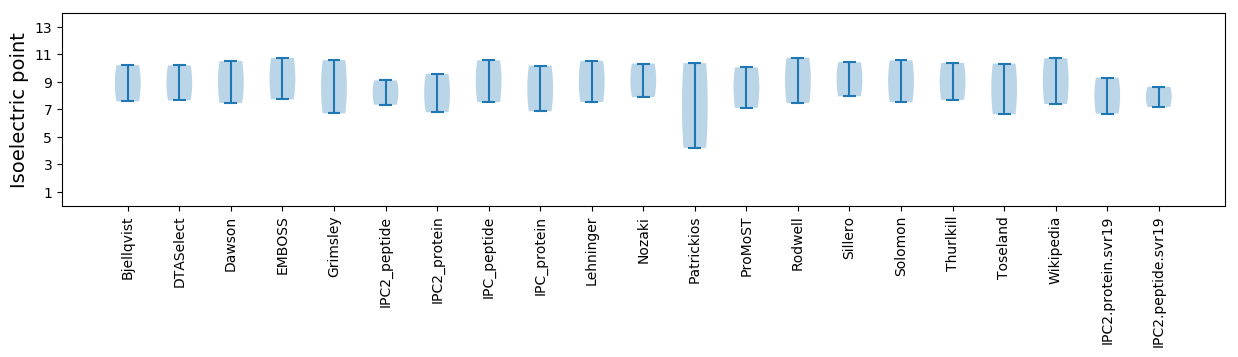
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGS1|A0A220IGS1_9CIRC Capsid protein OS=Giant panda circovirus 1 OX=2016456 PE=4 SV=1
MM1 pKa = 7.45VIGLMSLRR9 pKa = 11.84PRR11 pKa = 11.84YY12 pKa = 9.37RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SITPSDD21 pKa = 3.57VASFAWRR28 pKa = 11.84NRR30 pKa = 11.84RR31 pKa = 11.84TVHH34 pKa = 6.47NGILRR39 pKa = 11.84SGEE42 pKa = 3.85WVRR45 pKa = 11.84RR46 pKa = 11.84NAARR50 pKa = 11.84RR51 pKa = 11.84SSTSTVSTGGSRR63 pKa = 11.84RR64 pKa = 11.84SSLLSTGAASVKK76 pKa = 9.21RR77 pKa = 11.84TQLQGGATEE86 pKa = 3.79RR87 pKa = 11.84TARR90 pKa = 11.84VKK92 pKa = 10.55RR93 pKa = 11.84YY94 pKa = 9.18GSKK97 pKa = 10.14LKK99 pKa = 10.6KK100 pKa = 8.93IGRR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84VKK107 pKa = 10.24VSRR110 pKa = 11.84KK111 pKa = 8.98LRR113 pKa = 11.84AKK115 pKa = 10.15IIEE118 pKa = 4.14VTDD121 pKa = 3.71GNKK124 pKa = 9.3IQGYY128 pKa = 7.98YY129 pKa = 8.77QDD131 pKa = 3.95NRR133 pKa = 11.84IDD135 pKa = 3.72VLDD138 pKa = 4.58PGALGGRR145 pKa = 11.84QNVEE149 pKa = 3.44IFPNRR154 pKa = 11.84GGAASGYY161 pKa = 7.71MFNYY165 pKa = 10.11DD166 pKa = 3.44RR167 pKa = 11.84VLHH170 pKa = 5.7AASRR174 pKa = 11.84LWNTKK179 pKa = 9.89AAVQNPVYY187 pKa = 10.83SDD189 pKa = 3.77ALNFSPNDD197 pKa = 3.46TVINVEE203 pKa = 4.2KK204 pKa = 10.56QWWTFEE210 pKa = 3.86LRR212 pKa = 11.84NNSTRR217 pKa = 11.84TATLKK222 pKa = 9.24FCKK225 pKa = 9.5MRR227 pKa = 11.84MKK229 pKa = 10.5RR230 pKa = 11.84GCAQTLPYY238 pKa = 8.89DD239 pKa = 3.39TWVNGLAQMTTSGEE253 pKa = 4.27LVGGATINTMFTMPTLSTQFKK274 pKa = 8.26TAYY277 pKa = 8.13KK278 pKa = 10.36LEE280 pKa = 4.4VIKK283 pKa = 11.09VVLEE287 pKa = 4.11PGQSYY292 pKa = 8.14TFNIEE297 pKa = 3.94GPAMTYY303 pKa = 10.1HH304 pKa = 6.2GQDD307 pKa = 3.04FFDD310 pKa = 3.76NGTYY314 pKa = 10.23RR315 pKa = 11.84PVQKK319 pKa = 10.34QDD321 pKa = 2.93TLGFVIEE328 pKa = 4.35HH329 pKa = 6.27VDD331 pKa = 3.38MVGTHH336 pKa = 5.64ATSEE340 pKa = 4.36GAAGNAGFAPDD351 pKa = 3.49TTEE354 pKa = 4.41ANQAQEE360 pKa = 4.01RR361 pKa = 11.84VYY363 pKa = 10.33MRR365 pKa = 11.84STYY368 pKa = 9.97HH369 pKa = 6.49CRR371 pKa = 11.84LNMPEE376 pKa = 4.0KK377 pKa = 10.84VGGISTAAASIFQNGNRR394 pKa = 11.84VRR396 pKa = 11.84RR397 pKa = 11.84TCVDD401 pKa = 2.73EE402 pKa = 5.1FMPTTFATTMHH413 pKa = 6.62RR414 pKa = 11.84RR415 pKa = 11.84DD416 pKa = 3.58EE417 pKa = 4.33EE418 pKa = 4.44NPVVDD423 pKa = 4.14MNN425 pKa = 4.32
MM1 pKa = 7.45VIGLMSLRR9 pKa = 11.84PRR11 pKa = 11.84YY12 pKa = 9.37RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SITPSDD21 pKa = 3.57VASFAWRR28 pKa = 11.84NRR30 pKa = 11.84RR31 pKa = 11.84TVHH34 pKa = 6.47NGILRR39 pKa = 11.84SGEE42 pKa = 3.85WVRR45 pKa = 11.84RR46 pKa = 11.84NAARR50 pKa = 11.84RR51 pKa = 11.84SSTSTVSTGGSRR63 pKa = 11.84RR64 pKa = 11.84SSLLSTGAASVKK76 pKa = 9.21RR77 pKa = 11.84TQLQGGATEE86 pKa = 3.79RR87 pKa = 11.84TARR90 pKa = 11.84VKK92 pKa = 10.55RR93 pKa = 11.84YY94 pKa = 9.18GSKK97 pKa = 10.14LKK99 pKa = 10.6KK100 pKa = 8.93IGRR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84VKK107 pKa = 10.24VSRR110 pKa = 11.84KK111 pKa = 8.98LRR113 pKa = 11.84AKK115 pKa = 10.15IIEE118 pKa = 4.14VTDD121 pKa = 3.71GNKK124 pKa = 9.3IQGYY128 pKa = 7.98YY129 pKa = 8.77QDD131 pKa = 3.95NRR133 pKa = 11.84IDD135 pKa = 3.72VLDD138 pKa = 4.58PGALGGRR145 pKa = 11.84QNVEE149 pKa = 3.44IFPNRR154 pKa = 11.84GGAASGYY161 pKa = 7.71MFNYY165 pKa = 10.11DD166 pKa = 3.44RR167 pKa = 11.84VLHH170 pKa = 5.7AASRR174 pKa = 11.84LWNTKK179 pKa = 9.89AAVQNPVYY187 pKa = 10.83SDD189 pKa = 3.77ALNFSPNDD197 pKa = 3.46TVINVEE203 pKa = 4.2KK204 pKa = 10.56QWWTFEE210 pKa = 3.86LRR212 pKa = 11.84NNSTRR217 pKa = 11.84TATLKK222 pKa = 9.24FCKK225 pKa = 9.5MRR227 pKa = 11.84MKK229 pKa = 10.5RR230 pKa = 11.84GCAQTLPYY238 pKa = 8.89DD239 pKa = 3.39TWVNGLAQMTTSGEE253 pKa = 4.27LVGGATINTMFTMPTLSTQFKK274 pKa = 8.26TAYY277 pKa = 8.13KK278 pKa = 10.36LEE280 pKa = 4.4VIKK283 pKa = 11.09VVLEE287 pKa = 4.11PGQSYY292 pKa = 8.14TFNIEE297 pKa = 3.94GPAMTYY303 pKa = 10.1HH304 pKa = 6.2GQDD307 pKa = 3.04FFDD310 pKa = 3.76NGTYY314 pKa = 10.23RR315 pKa = 11.84PVQKK319 pKa = 10.34QDD321 pKa = 2.93TLGFVIEE328 pKa = 4.35HH329 pKa = 6.27VDD331 pKa = 3.38MVGTHH336 pKa = 5.64ATSEE340 pKa = 4.36GAAGNAGFAPDD351 pKa = 3.49TTEE354 pKa = 4.41ANQAQEE360 pKa = 4.01RR361 pKa = 11.84VYY363 pKa = 10.33MRR365 pKa = 11.84STYY368 pKa = 9.97HH369 pKa = 6.49CRR371 pKa = 11.84LNMPEE376 pKa = 4.0KK377 pKa = 10.84VGGISTAAASIFQNGNRR394 pKa = 11.84VRR396 pKa = 11.84RR397 pKa = 11.84TCVDD401 pKa = 2.73EE402 pKa = 5.1FMPTTFATTMHH413 pKa = 6.62RR414 pKa = 11.84RR415 pKa = 11.84DD416 pKa = 3.58EE417 pKa = 4.33EE418 pKa = 4.44NPVVDD423 pKa = 4.14MNN425 pKa = 4.32
Molecular weight: 47.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
713 |
288 |
425 |
356.5 |
40.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.732 ± 1.102 | 1.683 ± 0.645 |
4.909 ± 0.79 | 5.189 ± 0.829 |
3.506 ± 0.225 | 7.854 ± 0.331 |
1.823 ± 0.153 | 3.927 ± 0.141 |
4.067 ± 0.35 | 5.891 ± 0.416 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.525 ± 0.873 | 4.909 ± 1.05 |
3.787 ± 0.224 | 4.208 ± 0.18 |
9.818 ± 0.056 | 7.293 ± 0.612 |
8.135 ± 1.314 | 7.854 ± 0.282 |
2.244 ± 0.723 | 3.647 ± 0.306 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
