
Halobiforma lacisalsi AJ5
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Halobiforma; Halobiforma lacisalsi
Average proteome isoelectric point is 4.79
Get precalculated fractions of proteins
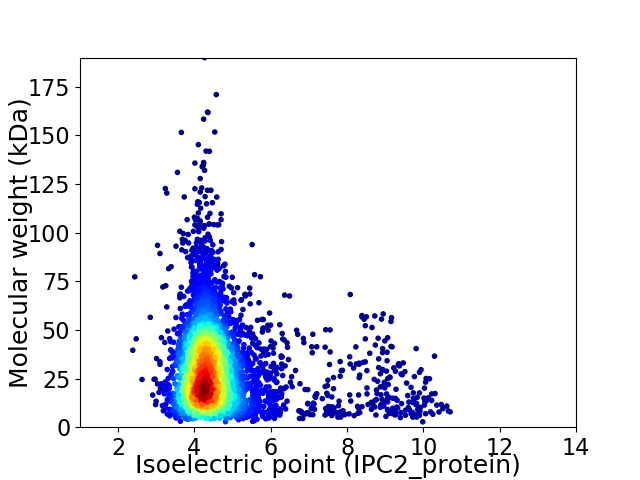
Virtual 2D-PAGE plot for 4151 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
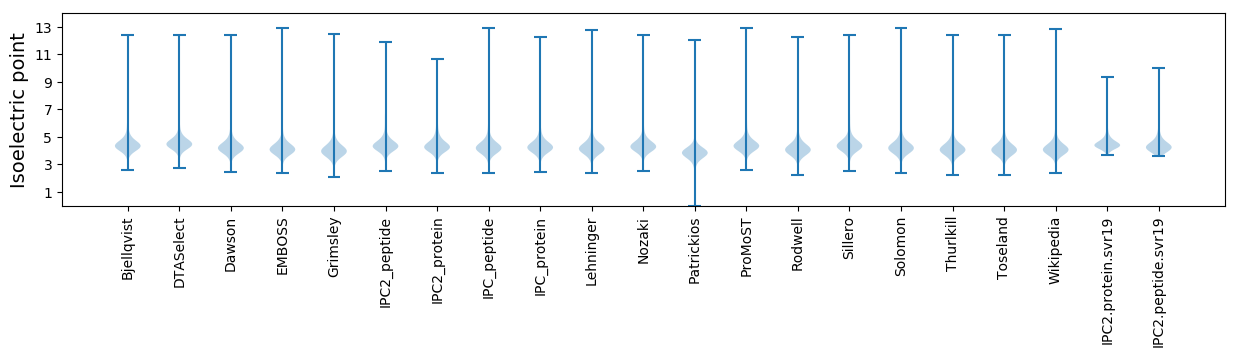
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0L9E0|M0L9E0_9EURY Transcription regulator AsnC-type-like protein OS=Halobiforma lacisalsi AJ5 OX=358396 GN=C445_16649 PE=4 SV=1
MM1 pKa = 7.71PPDD4 pKa = 4.77DD5 pKa = 5.18SPTSDD10 pKa = 2.82RR11 pKa = 11.84RR12 pKa = 11.84SVLKK16 pKa = 10.69AAGALGAFLGSSGVAAADD34 pKa = 3.75GDD36 pKa = 4.28DD37 pKa = 4.05EE38 pKa = 6.67ADD40 pKa = 3.45AGATEE45 pKa = 4.47LLVGVSPDD53 pKa = 3.31VSDD56 pKa = 3.87VEE58 pKa = 4.6SAVDD62 pKa = 3.45SSIVDD67 pKa = 3.63GQVVHH72 pKa = 6.2TNEE75 pKa = 4.84AIHH78 pKa = 6.05YY79 pKa = 7.12ATIEE83 pKa = 4.17FPEE86 pKa = 4.15NTPEE90 pKa = 3.87EE91 pKa = 4.31AKK93 pKa = 9.87TAIEE97 pKa = 4.28DD98 pKa = 3.57ALASVDD104 pKa = 3.57EE105 pKa = 4.29IEE107 pKa = 4.39YY108 pKa = 10.4VEE110 pKa = 4.34EE111 pKa = 3.95NATIEE116 pKa = 4.62SFAAPNDD123 pKa = 3.91PYY125 pKa = 11.03YY126 pKa = 10.98GSQHH130 pKa = 6.72APQQVNCEE138 pKa = 4.2GAWDD142 pKa = 3.89EE143 pKa = 4.46TFGDD147 pKa = 3.78SDD149 pKa = 3.93VTISIVDD156 pKa = 3.16TGTAYY161 pKa = 10.55DD162 pKa = 4.16HH163 pKa = 7.17EE164 pKa = 4.51NLAEE168 pKa = 4.18NVDD171 pKa = 3.72DD172 pKa = 5.52RR173 pKa = 11.84IGEE176 pKa = 4.24VFVGGGSDD184 pKa = 3.64PYY186 pKa = 10.21PRR188 pKa = 11.84SGSEE192 pKa = 3.54THH194 pKa = 5.47GTMVSGAAVGATDD207 pKa = 4.23NGTGHH212 pKa = 7.29AGVSNCSMLSARR224 pKa = 11.84ALDD227 pKa = 3.78ASGTGSLADD236 pKa = 3.82IADD239 pKa = 4.01AVQWSADD246 pKa = 3.37QGVDD250 pKa = 4.49VINLSLGSSSDD261 pKa = 2.68WHH263 pKa = 5.63TLRR266 pKa = 11.84NACQYY271 pKa = 11.39AYY273 pKa = 10.5DD274 pKa = 3.86QGCLLVAAAGNSGGSVAYY292 pKa = 8.75PAVYY296 pKa = 10.35DD297 pKa = 3.73SVIAVSALDD306 pKa = 3.39SHH308 pKa = 7.25DD309 pKa = 3.8RR310 pKa = 11.84LASFSNRR317 pKa = 11.84GSEE320 pKa = 4.35IEE322 pKa = 4.02LAAPGTSIVTTTLNDD337 pKa = 3.87GYY339 pKa = 9.23TRR341 pKa = 11.84ASGTSIASPIVAGVAGLVLSEE362 pKa = 4.31YY363 pKa = 10.13PDD365 pKa = 4.3LDD367 pKa = 3.69VEE369 pKa = 4.47TLRR372 pKa = 11.84KK373 pKa = 9.47HH374 pKa = 6.27LRR376 pKa = 11.84QTATDD381 pKa = 3.55VGLSAAAQGYY391 pKa = 9.48GRR393 pKa = 11.84VDD395 pKa = 2.94ADD397 pKa = 3.46AAVNTVPDD405 pKa = 4.51GYY407 pKa = 10.96EE408 pKa = 4.16PEE410 pKa = 4.24NPDD413 pKa = 4.08YY414 pKa = 11.61GDD416 pKa = 5.42DD417 pKa = 5.12DD418 pKa = 5.78DD419 pKa = 6.33EE420 pKa = 6.22EE421 pKa = 7.14DD422 pKa = 3.72DD423 pKa = 4.55TEE425 pKa = 5.66DD426 pKa = 4.01PGIDD430 pKa = 3.72EE431 pKa = 4.41VWNDD435 pKa = 3.46DD436 pKa = 3.54EE437 pKa = 5.22SNHH440 pKa = 5.8VEE442 pKa = 4.65LEE444 pKa = 4.12GEE446 pKa = 4.15SLDD449 pKa = 3.86RR450 pKa = 11.84VAEE453 pKa = 3.85YY454 pKa = 10.28RR455 pKa = 11.84IEE457 pKa = 4.23GRR459 pKa = 11.84GTAEE463 pKa = 3.98PGEE466 pKa = 4.29NANTDD471 pKa = 3.28PDD473 pKa = 3.93DD474 pKa = 4.41PYY476 pKa = 11.7RR477 pKa = 11.84DD478 pKa = 3.23IATTDD483 pKa = 3.16GEE485 pKa = 4.46EE486 pKa = 4.28FVVEE490 pKa = 5.18GYY492 pKa = 10.79LGGYY496 pKa = 9.47VDD498 pKa = 5.47DD499 pKa = 4.26FHH501 pKa = 6.95ITGAVTDD508 pKa = 3.79VEE510 pKa = 4.84TNVDD514 pKa = 3.48LTAVVNGHH522 pKa = 6.72AFDD525 pKa = 4.86LRR527 pKa = 11.84DD528 pKa = 3.64LEE530 pKa = 5.7GVGDD534 pKa = 4.1WEE536 pKa = 5.64ADD538 pKa = 3.6DD539 pKa = 6.33GEE541 pKa = 4.51PQCGDD546 pKa = 3.36EE547 pKa = 4.46TVTASAEE554 pKa = 4.2GSLSGRR560 pKa = 11.84WWGGTDD566 pKa = 2.9SYY568 pKa = 11.35TYY570 pKa = 10.59SLRR573 pKa = 11.84TADD576 pKa = 3.68PCSATVALDD585 pKa = 4.07GPDD588 pKa = 3.51GADD591 pKa = 3.09FDD593 pKa = 6.05LYY595 pKa = 10.21VTLDD599 pKa = 3.27GRR601 pKa = 11.84SPSRR605 pKa = 11.84WDD607 pKa = 3.27YY608 pKa = 11.83DD609 pKa = 3.64EE610 pKa = 6.18SSDD613 pKa = 4.58GSEE616 pKa = 3.85SDD618 pKa = 3.32EE619 pKa = 5.02SITVDD624 pKa = 4.21LEE626 pKa = 4.04GDD628 pKa = 3.47EE629 pKa = 4.27EE630 pKa = 5.04LRR632 pKa = 11.84LQVHH636 pKa = 6.76ANSGSGEE643 pKa = 4.14YY644 pKa = 10.25EE645 pKa = 3.88VTVEE649 pKa = 3.86EE650 pKa = 4.63RR651 pKa = 11.84GRR653 pKa = 3.57
MM1 pKa = 7.71PPDD4 pKa = 4.77DD5 pKa = 5.18SPTSDD10 pKa = 2.82RR11 pKa = 11.84RR12 pKa = 11.84SVLKK16 pKa = 10.69AAGALGAFLGSSGVAAADD34 pKa = 3.75GDD36 pKa = 4.28DD37 pKa = 4.05EE38 pKa = 6.67ADD40 pKa = 3.45AGATEE45 pKa = 4.47LLVGVSPDD53 pKa = 3.31VSDD56 pKa = 3.87VEE58 pKa = 4.6SAVDD62 pKa = 3.45SSIVDD67 pKa = 3.63GQVVHH72 pKa = 6.2TNEE75 pKa = 4.84AIHH78 pKa = 6.05YY79 pKa = 7.12ATIEE83 pKa = 4.17FPEE86 pKa = 4.15NTPEE90 pKa = 3.87EE91 pKa = 4.31AKK93 pKa = 9.87TAIEE97 pKa = 4.28DD98 pKa = 3.57ALASVDD104 pKa = 3.57EE105 pKa = 4.29IEE107 pKa = 4.39YY108 pKa = 10.4VEE110 pKa = 4.34EE111 pKa = 3.95NATIEE116 pKa = 4.62SFAAPNDD123 pKa = 3.91PYY125 pKa = 11.03YY126 pKa = 10.98GSQHH130 pKa = 6.72APQQVNCEE138 pKa = 4.2GAWDD142 pKa = 3.89EE143 pKa = 4.46TFGDD147 pKa = 3.78SDD149 pKa = 3.93VTISIVDD156 pKa = 3.16TGTAYY161 pKa = 10.55DD162 pKa = 4.16HH163 pKa = 7.17EE164 pKa = 4.51NLAEE168 pKa = 4.18NVDD171 pKa = 3.72DD172 pKa = 5.52RR173 pKa = 11.84IGEE176 pKa = 4.24VFVGGGSDD184 pKa = 3.64PYY186 pKa = 10.21PRR188 pKa = 11.84SGSEE192 pKa = 3.54THH194 pKa = 5.47GTMVSGAAVGATDD207 pKa = 4.23NGTGHH212 pKa = 7.29AGVSNCSMLSARR224 pKa = 11.84ALDD227 pKa = 3.78ASGTGSLADD236 pKa = 3.82IADD239 pKa = 4.01AVQWSADD246 pKa = 3.37QGVDD250 pKa = 4.49VINLSLGSSSDD261 pKa = 2.68WHH263 pKa = 5.63TLRR266 pKa = 11.84NACQYY271 pKa = 11.39AYY273 pKa = 10.5DD274 pKa = 3.86QGCLLVAAAGNSGGSVAYY292 pKa = 8.75PAVYY296 pKa = 10.35DD297 pKa = 3.73SVIAVSALDD306 pKa = 3.39SHH308 pKa = 7.25DD309 pKa = 3.8RR310 pKa = 11.84LASFSNRR317 pKa = 11.84GSEE320 pKa = 4.35IEE322 pKa = 4.02LAAPGTSIVTTTLNDD337 pKa = 3.87GYY339 pKa = 9.23TRR341 pKa = 11.84ASGTSIASPIVAGVAGLVLSEE362 pKa = 4.31YY363 pKa = 10.13PDD365 pKa = 4.3LDD367 pKa = 3.69VEE369 pKa = 4.47TLRR372 pKa = 11.84KK373 pKa = 9.47HH374 pKa = 6.27LRR376 pKa = 11.84QTATDD381 pKa = 3.55VGLSAAAQGYY391 pKa = 9.48GRR393 pKa = 11.84VDD395 pKa = 2.94ADD397 pKa = 3.46AAVNTVPDD405 pKa = 4.51GYY407 pKa = 10.96EE408 pKa = 4.16PEE410 pKa = 4.24NPDD413 pKa = 4.08YY414 pKa = 11.61GDD416 pKa = 5.42DD417 pKa = 5.12DD418 pKa = 5.78DD419 pKa = 6.33EE420 pKa = 6.22EE421 pKa = 7.14DD422 pKa = 3.72DD423 pKa = 4.55TEE425 pKa = 5.66DD426 pKa = 4.01PGIDD430 pKa = 3.72EE431 pKa = 4.41VWNDD435 pKa = 3.46DD436 pKa = 3.54EE437 pKa = 5.22SNHH440 pKa = 5.8VEE442 pKa = 4.65LEE444 pKa = 4.12GEE446 pKa = 4.15SLDD449 pKa = 3.86RR450 pKa = 11.84VAEE453 pKa = 3.85YY454 pKa = 10.28RR455 pKa = 11.84IEE457 pKa = 4.23GRR459 pKa = 11.84GTAEE463 pKa = 3.98PGEE466 pKa = 4.29NANTDD471 pKa = 3.28PDD473 pKa = 3.93DD474 pKa = 4.41PYY476 pKa = 11.7RR477 pKa = 11.84DD478 pKa = 3.23IATTDD483 pKa = 3.16GEE485 pKa = 4.46EE486 pKa = 4.28FVVEE490 pKa = 5.18GYY492 pKa = 10.79LGGYY496 pKa = 9.47VDD498 pKa = 5.47DD499 pKa = 4.26FHH501 pKa = 6.95ITGAVTDD508 pKa = 3.79VEE510 pKa = 4.84TNVDD514 pKa = 3.48LTAVVNGHH522 pKa = 6.72AFDD525 pKa = 4.86LRR527 pKa = 11.84DD528 pKa = 3.64LEE530 pKa = 5.7GVGDD534 pKa = 4.1WEE536 pKa = 5.64ADD538 pKa = 3.6DD539 pKa = 6.33GEE541 pKa = 4.51PQCGDD546 pKa = 3.36EE547 pKa = 4.46TVTASAEE554 pKa = 4.2GSLSGRR560 pKa = 11.84WWGGTDD566 pKa = 2.9SYY568 pKa = 11.35TYY570 pKa = 10.59SLRR573 pKa = 11.84TADD576 pKa = 3.68PCSATVALDD585 pKa = 4.07GPDD588 pKa = 3.51GADD591 pKa = 3.09FDD593 pKa = 6.05LYY595 pKa = 10.21VTLDD599 pKa = 3.27GRR601 pKa = 11.84SPSRR605 pKa = 11.84WDD607 pKa = 3.27YY608 pKa = 11.83DD609 pKa = 3.64EE610 pKa = 6.18SSDD613 pKa = 4.58GSEE616 pKa = 3.85SDD618 pKa = 3.32EE619 pKa = 5.02SITVDD624 pKa = 4.21LEE626 pKa = 4.04GDD628 pKa = 3.47EE629 pKa = 4.27EE630 pKa = 5.04LRR632 pKa = 11.84LQVHH636 pKa = 6.76ANSGSGEE643 pKa = 4.14YY644 pKa = 10.25EE645 pKa = 3.88VTVEE649 pKa = 3.86EE650 pKa = 4.63RR651 pKa = 11.84GRR653 pKa = 3.57
Molecular weight: 68.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0LTS9|M0LTS9_9EURY 2-keto-3-deoxygluconate kinase OS=Halobiforma lacisalsi AJ5 OX=358396 GN=C445_01956 PE=4 SV=1
MM1 pKa = 7.34AADD4 pKa = 4.16RR5 pKa = 11.84GYY7 pKa = 11.06DD8 pKa = 3.32DD9 pKa = 4.22KK10 pKa = 11.8AFRR13 pKa = 11.84DD14 pKa = 3.75EE15 pKa = 4.49LRR17 pKa = 11.84ANGIRR22 pKa = 11.84PLIKK26 pKa = 10.2HH27 pKa = 6.53RR28 pKa = 11.84IYY30 pKa = 9.95WALDD34 pKa = 3.19HH35 pKa = 6.59AHH37 pKa = 6.85NARR40 pKa = 11.84MDD42 pKa = 3.65RR43 pKa = 11.84DD44 pKa = 3.47RR45 pKa = 11.84YY46 pKa = 8.01NRR48 pKa = 11.84RR49 pKa = 11.84WMVEE53 pKa = 4.13TVFSSLKK60 pKa = 8.84RR61 pKa = 11.84TLGSAVRR68 pKa = 11.84ARR70 pKa = 11.84SWHH73 pKa = 5.61LEE75 pKa = 3.59FRR77 pKa = 11.84EE78 pKa = 4.1MVLKK82 pKa = 10.7CAVYY86 pKa = 9.85NLRR89 pKa = 11.84RR90 pKa = 11.84TVRR93 pKa = 11.84YY94 pKa = 8.29PP95 pKa = 2.95
MM1 pKa = 7.34AADD4 pKa = 4.16RR5 pKa = 11.84GYY7 pKa = 11.06DD8 pKa = 3.32DD9 pKa = 4.22KK10 pKa = 11.8AFRR13 pKa = 11.84DD14 pKa = 3.75EE15 pKa = 4.49LRR17 pKa = 11.84ANGIRR22 pKa = 11.84PLIKK26 pKa = 10.2HH27 pKa = 6.53RR28 pKa = 11.84IYY30 pKa = 9.95WALDD34 pKa = 3.19HH35 pKa = 6.59AHH37 pKa = 6.85NARR40 pKa = 11.84MDD42 pKa = 3.65RR43 pKa = 11.84DD44 pKa = 3.47RR45 pKa = 11.84YY46 pKa = 8.01NRR48 pKa = 11.84RR49 pKa = 11.84WMVEE53 pKa = 4.13TVFSSLKK60 pKa = 8.84RR61 pKa = 11.84TLGSAVRR68 pKa = 11.84ARR70 pKa = 11.84SWHH73 pKa = 5.61LEE75 pKa = 3.59FRR77 pKa = 11.84EE78 pKa = 4.1MVLKK82 pKa = 10.7CAVYY86 pKa = 9.85NLRR89 pKa = 11.84RR90 pKa = 11.84TVRR93 pKa = 11.84YY94 pKa = 8.29PP95 pKa = 2.95
Molecular weight: 11.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212350 |
24 |
1709 |
292.1 |
31.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.605 ± 0.055 | 0.72 ± 0.013 |
8.783 ± 0.053 | 9.646 ± 0.062 |
3.21 ± 0.027 | 8.569 ± 0.039 |
1.91 ± 0.019 | 4.074 ± 0.028 |
1.65 ± 0.024 | 8.855 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.654 ± 0.016 | 2.177 ± 0.019 |
4.743 ± 0.027 | 2.219 ± 0.025 |
6.798 ± 0.04 | 5.415 ± 0.025 |
6.295 ± 0.032 | 8.881 ± 0.041 |
1.112 ± 0.016 | 2.681 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
