
Avon-Heathcote Estuary associated circular virus 18
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae; Avon-Heathcote Estuary associated circular virus 17
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
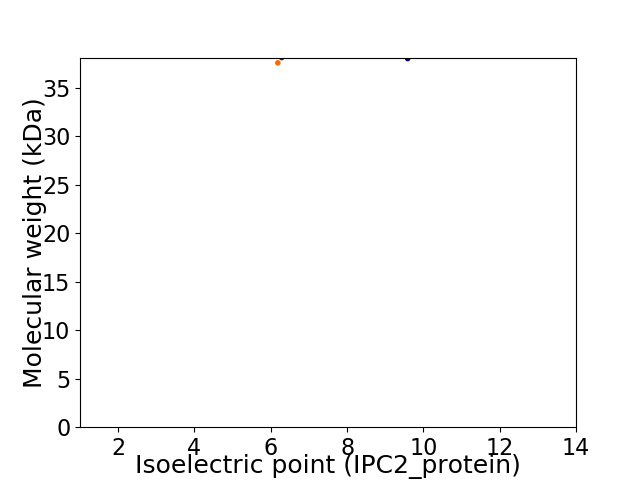
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5I9U1|A0A0C5I9U1_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 18 OX=1618241 PE=4 SV=1
MM1 pKa = 7.39TARR4 pKa = 11.84NWVFTINNHH13 pKa = 5.94FSIDD17 pKa = 3.89LEE19 pKa = 4.24EE20 pKa = 4.17EE21 pKa = 3.86TWNNNLKK28 pKa = 10.67LLIAVKK34 pKa = 10.16EE35 pKa = 4.11VGEE38 pKa = 4.14QGTEE42 pKa = 3.73HH43 pKa = 6.09LQGYY47 pKa = 9.67LEE49 pKa = 5.39LIQPRR54 pKa = 11.84RR55 pKa = 11.84FSWVKK60 pKa = 9.74EE61 pKa = 3.94RR62 pKa = 11.84LPLAHH67 pKa = 6.77LEE69 pKa = 4.01KK70 pKa = 10.64RR71 pKa = 11.84RR72 pKa = 11.84GSKK75 pKa = 9.68KK76 pKa = 9.84QAIQYY81 pKa = 8.36CLKK84 pKa = 9.93TISLPSQQCSSPSNTSDD101 pKa = 3.53TSPQDD106 pKa = 3.17VDD108 pKa = 3.53STIPLTIIYY117 pKa = 9.0PPDD120 pKa = 3.45MNLSDD125 pKa = 5.66LYY127 pKa = 11.53ADD129 pKa = 3.75YY130 pKa = 11.06CRR132 pKa = 11.84SKK134 pKa = 8.78QTKK137 pKa = 9.35RR138 pKa = 11.84QILLAEE144 pKa = 4.37VRR146 pKa = 11.84QKK148 pKa = 11.09LSEE151 pKa = 4.23GVDD154 pKa = 3.46DD155 pKa = 3.91LTIANEE161 pKa = 4.11YY162 pKa = 9.85FDD164 pKa = 4.34VWVSNYY170 pKa = 10.11RR171 pKa = 11.84AFEE174 pKa = 4.1RR175 pKa = 11.84YY176 pKa = 7.39RR177 pKa = 11.84TLITPPRR184 pKa = 11.84NHH186 pKa = 6.44EE187 pKa = 4.1VNVIVIQGPTGTGKK201 pKa = 10.7SKK203 pKa = 9.63WCLDD207 pKa = 4.09NYY209 pKa = 10.08PNAYY213 pKa = 8.18WKK215 pKa = 10.05QRR217 pKa = 11.84SNWWDD222 pKa = 3.77GYY224 pKa = 10.33SGHH227 pKa = 6.99DD228 pKa = 3.28TVIIDD233 pKa = 4.6EE234 pKa = 4.88FYY236 pKa = 10.81GWLPFDD242 pKa = 4.26LLLRR246 pKa = 11.84VCDD249 pKa = 4.56RR250 pKa = 11.84YY251 pKa = 10.46PLLVEE256 pKa = 4.41SKK258 pKa = 10.15GGQIQFVAKK267 pKa = 9.65TIIITSNSVPSSWYY281 pKa = 8.66RR282 pKa = 11.84NSYY285 pKa = 9.31FDD287 pKa = 3.29SFVRR291 pKa = 11.84RR292 pKa = 11.84VSKK295 pKa = 10.47WMVFPIWGDD304 pKa = 3.29QLSFTDD310 pKa = 3.83YY311 pKa = 11.26SEE313 pKa = 5.89AIAHH317 pKa = 6.14FVNNYY322 pKa = 8.86
MM1 pKa = 7.39TARR4 pKa = 11.84NWVFTINNHH13 pKa = 5.94FSIDD17 pKa = 3.89LEE19 pKa = 4.24EE20 pKa = 4.17EE21 pKa = 3.86TWNNNLKK28 pKa = 10.67LLIAVKK34 pKa = 10.16EE35 pKa = 4.11VGEE38 pKa = 4.14QGTEE42 pKa = 3.73HH43 pKa = 6.09LQGYY47 pKa = 9.67LEE49 pKa = 5.39LIQPRR54 pKa = 11.84RR55 pKa = 11.84FSWVKK60 pKa = 9.74EE61 pKa = 3.94RR62 pKa = 11.84LPLAHH67 pKa = 6.77LEE69 pKa = 4.01KK70 pKa = 10.64RR71 pKa = 11.84RR72 pKa = 11.84GSKK75 pKa = 9.68KK76 pKa = 9.84QAIQYY81 pKa = 8.36CLKK84 pKa = 9.93TISLPSQQCSSPSNTSDD101 pKa = 3.53TSPQDD106 pKa = 3.17VDD108 pKa = 3.53STIPLTIIYY117 pKa = 9.0PPDD120 pKa = 3.45MNLSDD125 pKa = 5.66LYY127 pKa = 11.53ADD129 pKa = 3.75YY130 pKa = 11.06CRR132 pKa = 11.84SKK134 pKa = 8.78QTKK137 pKa = 9.35RR138 pKa = 11.84QILLAEE144 pKa = 4.37VRR146 pKa = 11.84QKK148 pKa = 11.09LSEE151 pKa = 4.23GVDD154 pKa = 3.46DD155 pKa = 3.91LTIANEE161 pKa = 4.11YY162 pKa = 9.85FDD164 pKa = 4.34VWVSNYY170 pKa = 10.11RR171 pKa = 11.84AFEE174 pKa = 4.1RR175 pKa = 11.84YY176 pKa = 7.39RR177 pKa = 11.84TLITPPRR184 pKa = 11.84NHH186 pKa = 6.44EE187 pKa = 4.1VNVIVIQGPTGTGKK201 pKa = 10.7SKK203 pKa = 9.63WCLDD207 pKa = 4.09NYY209 pKa = 10.08PNAYY213 pKa = 8.18WKK215 pKa = 10.05QRR217 pKa = 11.84SNWWDD222 pKa = 3.77GYY224 pKa = 10.33SGHH227 pKa = 6.99DD228 pKa = 3.28TVIIDD233 pKa = 4.6EE234 pKa = 4.88FYY236 pKa = 10.81GWLPFDD242 pKa = 4.26LLLRR246 pKa = 11.84VCDD249 pKa = 4.56RR250 pKa = 11.84YY251 pKa = 10.46PLLVEE256 pKa = 4.41SKK258 pKa = 10.15GGQIQFVAKK267 pKa = 9.65TIIITSNSVPSSWYY281 pKa = 8.66RR282 pKa = 11.84NSYY285 pKa = 9.31FDD287 pKa = 3.29SFVRR291 pKa = 11.84RR292 pKa = 11.84VSKK295 pKa = 10.47WMVFPIWGDD304 pKa = 3.29QLSFTDD310 pKa = 3.83YY311 pKa = 11.26SEE313 pKa = 5.89AIAHH317 pKa = 6.14FVNNYY322 pKa = 8.86
Molecular weight: 37.59 kDa
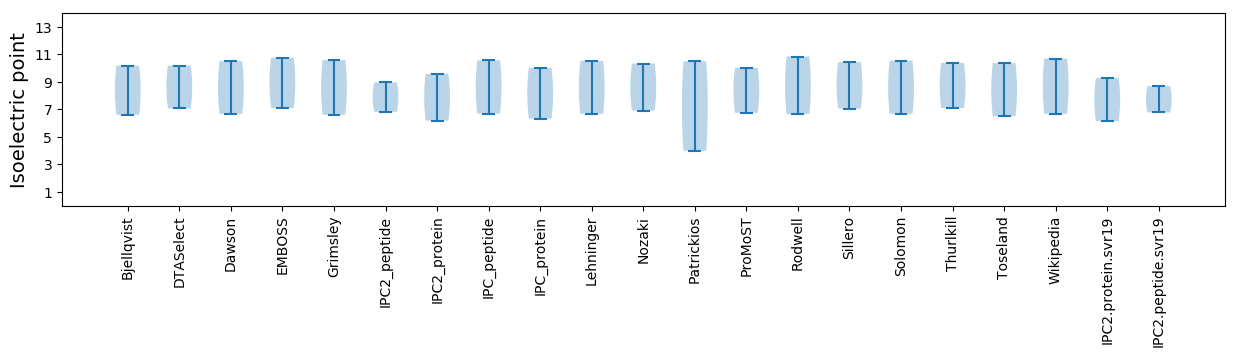
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5I9U1|A0A0C5I9U1_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 18 OX=1618241 PE=4 SV=1
MM1 pKa = 8.17PYY3 pKa = 9.98RR4 pKa = 11.84PKK6 pKa = 10.26PYY8 pKa = 9.88SRR10 pKa = 11.84SLVDD14 pKa = 2.84YY15 pKa = 10.56RR16 pKa = 11.84RR17 pKa = 11.84ASRR20 pKa = 11.84INYY23 pKa = 9.74RR24 pKa = 11.84IGARR28 pKa = 11.84LANRR32 pKa = 11.84VGMSFTKK39 pKa = 10.01TKK41 pKa = 10.5RR42 pKa = 11.84KK43 pKa = 9.36RR44 pKa = 11.84QTSGRR49 pKa = 11.84GVTFQRR55 pKa = 11.84DD56 pKa = 3.0QEE58 pKa = 4.3LIYY61 pKa = 10.33RR62 pKa = 11.84KK63 pKa = 9.76RR64 pKa = 11.84RR65 pKa = 11.84MPRR68 pKa = 11.84KK69 pKa = 9.4RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84WKK75 pKa = 9.22NFIRR79 pKa = 11.84KK80 pKa = 8.66SVAASEE86 pKa = 4.31KK87 pKa = 8.8TLGSRR92 pKa = 11.84TVVRR96 pKa = 11.84NQQFSFAVSGVNANSEE112 pKa = 4.53DD113 pKa = 3.63IVGQLALYY121 pKa = 9.81AVQNGGNEE129 pKa = 3.98YY130 pKa = 10.74LRR132 pKa = 11.84DD133 pKa = 3.73LNRR136 pKa = 11.84ICSDD140 pKa = 3.33TDD142 pKa = 3.24VSTSGKK148 pKa = 10.58LIFQSGVMDD157 pKa = 3.55ATITNISSSDD167 pKa = 3.42DD168 pKa = 3.38QDD170 pKa = 4.07RR171 pKa = 11.84KK172 pKa = 10.75DD173 pKa = 3.67VPIEE177 pKa = 3.57LDD179 pKa = 3.03IYY181 pKa = 10.37EE182 pKa = 4.14VSARR186 pKa = 11.84RR187 pKa = 11.84NFEE190 pKa = 3.9KK191 pKa = 10.58GTGDD195 pKa = 5.35KK196 pKa = 11.33GLLDD200 pKa = 4.14CFAEE204 pKa = 4.84GFLDD208 pKa = 4.3QGLLSTTGTDD218 pKa = 3.31TPLSLTTRR226 pKa = 11.84GSTPWDD232 pKa = 3.73CPQALSMYY240 pKa = 10.21RR241 pKa = 11.84LKK243 pKa = 10.22IWKK246 pKa = 6.87KK247 pKa = 7.45TKK249 pKa = 9.7YY250 pKa = 9.4RR251 pKa = 11.84LSGGQSLSYY260 pKa = 10.66QMRR263 pKa = 11.84DD264 pKa = 3.4PKK266 pKa = 10.48RR267 pKa = 11.84HH268 pKa = 4.78VMEE271 pKa = 4.74KK272 pKa = 10.52GYY274 pKa = 10.81VGEE277 pKa = 4.23LTGSNLPGVTRR288 pKa = 11.84WLILIAKK295 pKa = 7.95PIVGFNVNFNGIAEE309 pKa = 4.1IRR311 pKa = 11.84VGVTRR316 pKa = 11.84KK317 pKa = 9.85YY318 pKa = 10.59LYY320 pKa = 10.51KK321 pKa = 10.65LNEE324 pKa = 3.96RR325 pKa = 11.84NEE327 pKa = 4.35DD328 pKa = 3.17KK329 pKa = 10.99DD330 pKa = 3.95ALVTT334 pKa = 3.91
MM1 pKa = 8.17PYY3 pKa = 9.98RR4 pKa = 11.84PKK6 pKa = 10.26PYY8 pKa = 9.88SRR10 pKa = 11.84SLVDD14 pKa = 2.84YY15 pKa = 10.56RR16 pKa = 11.84RR17 pKa = 11.84ASRR20 pKa = 11.84INYY23 pKa = 9.74RR24 pKa = 11.84IGARR28 pKa = 11.84LANRR32 pKa = 11.84VGMSFTKK39 pKa = 10.01TKK41 pKa = 10.5RR42 pKa = 11.84KK43 pKa = 9.36RR44 pKa = 11.84QTSGRR49 pKa = 11.84GVTFQRR55 pKa = 11.84DD56 pKa = 3.0QEE58 pKa = 4.3LIYY61 pKa = 10.33RR62 pKa = 11.84KK63 pKa = 9.76RR64 pKa = 11.84RR65 pKa = 11.84MPRR68 pKa = 11.84KK69 pKa = 9.4RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84WKK75 pKa = 9.22NFIRR79 pKa = 11.84KK80 pKa = 8.66SVAASEE86 pKa = 4.31KK87 pKa = 8.8TLGSRR92 pKa = 11.84TVVRR96 pKa = 11.84NQQFSFAVSGVNANSEE112 pKa = 4.53DD113 pKa = 3.63IVGQLALYY121 pKa = 9.81AVQNGGNEE129 pKa = 3.98YY130 pKa = 10.74LRR132 pKa = 11.84DD133 pKa = 3.73LNRR136 pKa = 11.84ICSDD140 pKa = 3.33TDD142 pKa = 3.24VSTSGKK148 pKa = 10.58LIFQSGVMDD157 pKa = 3.55ATITNISSSDD167 pKa = 3.42DD168 pKa = 3.38QDD170 pKa = 4.07RR171 pKa = 11.84KK172 pKa = 10.75DD173 pKa = 3.67VPIEE177 pKa = 3.57LDD179 pKa = 3.03IYY181 pKa = 10.37EE182 pKa = 4.14VSARR186 pKa = 11.84RR187 pKa = 11.84NFEE190 pKa = 3.9KK191 pKa = 10.58GTGDD195 pKa = 5.35KK196 pKa = 11.33GLLDD200 pKa = 4.14CFAEE204 pKa = 4.84GFLDD208 pKa = 4.3QGLLSTTGTDD218 pKa = 3.31TPLSLTTRR226 pKa = 11.84GSTPWDD232 pKa = 3.73CPQALSMYY240 pKa = 10.21RR241 pKa = 11.84LKK243 pKa = 10.22IWKK246 pKa = 6.87KK247 pKa = 7.45TKK249 pKa = 9.7YY250 pKa = 9.4RR251 pKa = 11.84LSGGQSLSYY260 pKa = 10.66QMRR263 pKa = 11.84DD264 pKa = 3.4PKK266 pKa = 10.48RR267 pKa = 11.84HH268 pKa = 4.78VMEE271 pKa = 4.74KK272 pKa = 10.52GYY274 pKa = 10.81VGEE277 pKa = 4.23LTGSNLPGVTRR288 pKa = 11.84WLILIAKK295 pKa = 7.95PIVGFNVNFNGIAEE309 pKa = 4.1IRR311 pKa = 11.84VGVTRR316 pKa = 11.84KK317 pKa = 9.85YY318 pKa = 10.59LYY320 pKa = 10.51KK321 pKa = 10.65LNEE324 pKa = 3.96RR325 pKa = 11.84NEE327 pKa = 4.35DD328 pKa = 3.17KK329 pKa = 10.99DD330 pKa = 3.95ALVTT334 pKa = 3.91
Molecular weight: 38.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656 |
322 |
334 |
328.0 |
37.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.268 ± 0.376 | 1.22 ± 0.231 |
5.945 ± 0.031 | 4.573 ± 0.491 |
3.659 ± 0.263 | 6.25 ± 1.321 |
1.067 ± 0.553 | 5.945 ± 0.616 |
5.945 ± 0.678 | 8.537 ± 0.11 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.524 ± 0.412 | 5.488 ± 0.287 |
4.116 ± 0.592 | 4.421 ± 0.381 |
8.537 ± 1.831 | 8.384 ± 0.216 |
6.402 ± 0.348 | 6.555 ± 0.023 |
2.439 ± 0.894 | 4.726 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
