
Sewage-associated gemycircularvirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus sewopo2; Sewage derived gemykibivirus 2
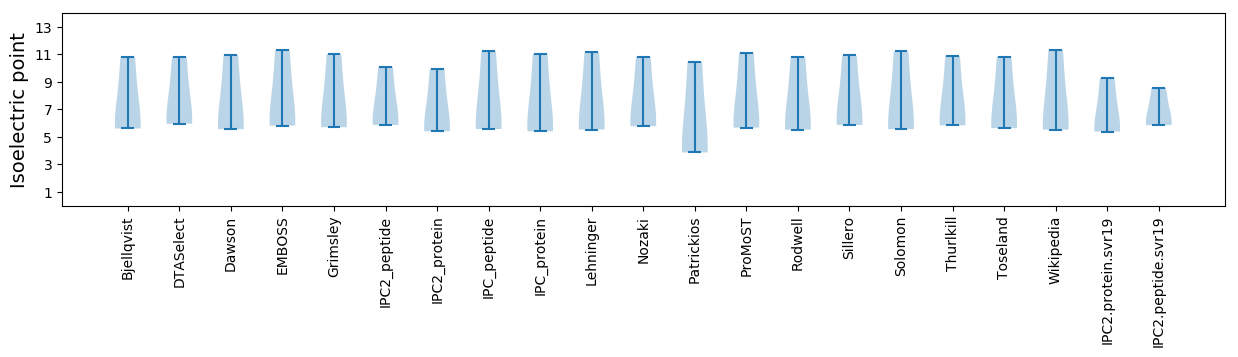
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
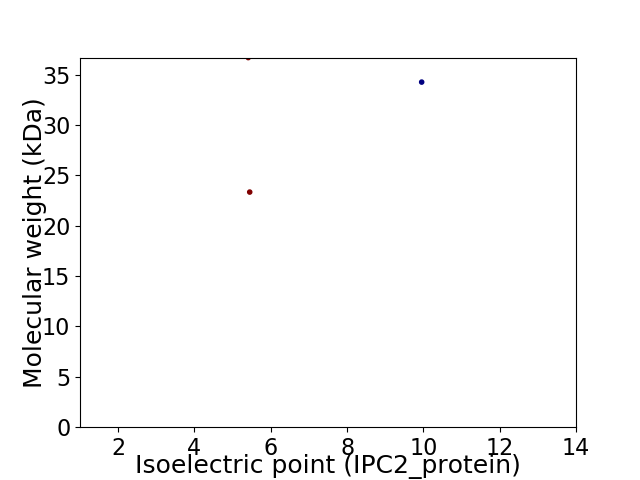
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
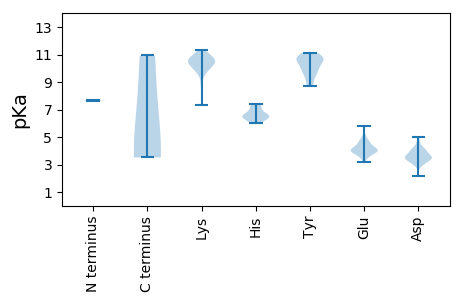
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7CL77|A0A0A7CL77_9VIRU Capsid protein OS=Sewage-associated gemycircularvirus 2 OX=1519407 PE=4 SV=1
MM1 pKa = 7.63PNAVTWKK8 pKa = 9.7LRR10 pKa = 11.84YY11 pKa = 10.17GIVTYY16 pKa = 9.59AQCSDD21 pKa = 3.65LDD23 pKa = 3.53PWKK26 pKa = 10.71VVDD29 pKa = 3.8MFATISAEE37 pKa = 4.06CIIGRR42 pKa = 11.84EE43 pKa = 3.95YY44 pKa = 11.1HH45 pKa = 6.53EE46 pKa = 5.79DD47 pKa = 3.46GGIHH51 pKa = 6.01LHH53 pKa = 6.45CFVDD57 pKa = 4.47FGRR60 pKa = 11.84IFRR63 pKa = 11.84SRR65 pKa = 11.84RR66 pKa = 11.84VDD68 pKa = 3.31VFDD71 pKa = 4.17VEE73 pKa = 4.89GRR75 pKa = 11.84HH76 pKa = 6.44PNVQHH81 pKa = 6.45VGRR84 pKa = 11.84TPWVAYY90 pKa = 9.42DD91 pKa = 3.46YY92 pKa = 10.52CIKK95 pKa = 10.8EE96 pKa = 4.07GDD98 pKa = 4.06VVAGAAEE105 pKa = 4.07RR106 pKa = 11.84PKK108 pKa = 10.43EE109 pKa = 3.94RR110 pKa = 11.84RR111 pKa = 11.84VGLSKK116 pKa = 10.44PDD118 pKa = 4.19EE119 pKa = 4.01IWTEE123 pKa = 3.62IMAATDD129 pKa = 3.53RR130 pKa = 11.84EE131 pKa = 4.63TFFYY135 pKa = 10.63LCQQLAPRR143 pKa = 11.84SLCLSFNSITAFAEE157 pKa = 3.47WKK159 pKa = 10.28YY160 pKa = 10.87RR161 pKa = 11.84VDD163 pKa = 3.58PEE165 pKa = 4.73PYY167 pKa = 8.73QHH169 pKa = 7.43PDD171 pKa = 3.32GIEE174 pKa = 3.77FLEE177 pKa = 4.32DD178 pKa = 2.99RR179 pKa = 11.84VGEE182 pKa = 3.87LHH184 pKa = 6.92RR185 pKa = 11.84WAEE188 pKa = 4.08ANLGLRR194 pKa = 11.84TGEE197 pKa = 4.05RR198 pKa = 11.84ARR200 pKa = 11.84SLVLIGPTRR209 pKa = 11.84VGKK212 pKa = 7.32TLWARR217 pKa = 11.84SLGNHH222 pKa = 7.12AYY224 pKa = 10.41FGGLFSLDD232 pKa = 3.43EE233 pKa = 4.11SLKK236 pKa = 9.9DD237 pKa = 3.17VEE239 pKa = 4.49YY240 pKa = 11.05AVFDD244 pKa = 4.09DD245 pKa = 4.2MMGGLEE251 pKa = 4.37FFHH254 pKa = 7.34SYY256 pKa = 10.65KK257 pKa = 10.45FWLGAQKK264 pKa = 10.36QFYY267 pKa = 8.95ATDD270 pKa = 3.38KK271 pKa = 10.64YY272 pKa = 10.71KK273 pKa = 10.99GKK275 pKa = 10.52RR276 pKa = 11.84LVDD279 pKa = 3.34WGRR282 pKa = 11.84PCIYY286 pKa = 10.01CHH288 pKa = 6.66NKK290 pKa = 10.08DD291 pKa = 3.57PRR293 pKa = 11.84LDD295 pKa = 3.67PKK297 pKa = 11.36ADD299 pKa = 3.67IDD301 pKa = 3.93WLEE304 pKa = 4.25GNCDD308 pKa = 4.82FITVEE313 pKa = 5.07DD314 pKa = 4.0SLLVV318 pKa = 3.6
MM1 pKa = 7.63PNAVTWKK8 pKa = 9.7LRR10 pKa = 11.84YY11 pKa = 10.17GIVTYY16 pKa = 9.59AQCSDD21 pKa = 3.65LDD23 pKa = 3.53PWKK26 pKa = 10.71VVDD29 pKa = 3.8MFATISAEE37 pKa = 4.06CIIGRR42 pKa = 11.84EE43 pKa = 3.95YY44 pKa = 11.1HH45 pKa = 6.53EE46 pKa = 5.79DD47 pKa = 3.46GGIHH51 pKa = 6.01LHH53 pKa = 6.45CFVDD57 pKa = 4.47FGRR60 pKa = 11.84IFRR63 pKa = 11.84SRR65 pKa = 11.84RR66 pKa = 11.84VDD68 pKa = 3.31VFDD71 pKa = 4.17VEE73 pKa = 4.89GRR75 pKa = 11.84HH76 pKa = 6.44PNVQHH81 pKa = 6.45VGRR84 pKa = 11.84TPWVAYY90 pKa = 9.42DD91 pKa = 3.46YY92 pKa = 10.52CIKK95 pKa = 10.8EE96 pKa = 4.07GDD98 pKa = 4.06VVAGAAEE105 pKa = 4.07RR106 pKa = 11.84PKK108 pKa = 10.43EE109 pKa = 3.94RR110 pKa = 11.84RR111 pKa = 11.84VGLSKK116 pKa = 10.44PDD118 pKa = 4.19EE119 pKa = 4.01IWTEE123 pKa = 3.62IMAATDD129 pKa = 3.53RR130 pKa = 11.84EE131 pKa = 4.63TFFYY135 pKa = 10.63LCQQLAPRR143 pKa = 11.84SLCLSFNSITAFAEE157 pKa = 3.47WKK159 pKa = 10.28YY160 pKa = 10.87RR161 pKa = 11.84VDD163 pKa = 3.58PEE165 pKa = 4.73PYY167 pKa = 8.73QHH169 pKa = 7.43PDD171 pKa = 3.32GIEE174 pKa = 3.77FLEE177 pKa = 4.32DD178 pKa = 2.99RR179 pKa = 11.84VGEE182 pKa = 3.87LHH184 pKa = 6.92RR185 pKa = 11.84WAEE188 pKa = 4.08ANLGLRR194 pKa = 11.84TGEE197 pKa = 4.05RR198 pKa = 11.84ARR200 pKa = 11.84SLVLIGPTRR209 pKa = 11.84VGKK212 pKa = 7.32TLWARR217 pKa = 11.84SLGNHH222 pKa = 7.12AYY224 pKa = 10.41FGGLFSLDD232 pKa = 3.43EE233 pKa = 4.11SLKK236 pKa = 9.9DD237 pKa = 3.17VEE239 pKa = 4.49YY240 pKa = 11.05AVFDD244 pKa = 4.09DD245 pKa = 4.2MMGGLEE251 pKa = 4.37FFHH254 pKa = 7.34SYY256 pKa = 10.65KK257 pKa = 10.45FWLGAQKK264 pKa = 10.36QFYY267 pKa = 8.95ATDD270 pKa = 3.38KK271 pKa = 10.64YY272 pKa = 10.71KK273 pKa = 10.99GKK275 pKa = 10.52RR276 pKa = 11.84LVDD279 pKa = 3.34WGRR282 pKa = 11.84PCIYY286 pKa = 10.01CHH288 pKa = 6.66NKK290 pKa = 10.08DD291 pKa = 3.57PRR293 pKa = 11.84LDD295 pKa = 3.67PKK297 pKa = 11.36ADD299 pKa = 3.67IDD301 pKa = 3.93WLEE304 pKa = 4.25GNCDD308 pKa = 4.82FITVEE313 pKa = 5.07DD314 pKa = 4.0SLLVV318 pKa = 3.6
Molecular weight: 36.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7CL77|A0A0A7CL77_9VIRU Capsid protein OS=Sewage-associated gemycircularvirus 2 OX=1519407 PE=4 SV=1
MM1 pKa = 7.66ALRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84LRR8 pKa = 11.84RR9 pKa = 11.84PTARR13 pKa = 11.84VRR15 pKa = 11.84PSRR18 pKa = 11.84ASGFRR23 pKa = 11.84RR24 pKa = 11.84VARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84FTRR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84IRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PMTNRR47 pKa = 11.84RR48 pKa = 11.84ILNVSSKK55 pKa = 10.86KK56 pKa = 10.48KK57 pKa = 9.95VDD59 pKa = 3.06TMMPLVIGNDD69 pKa = 3.85GVGTEE74 pKa = 4.49GPLLVTPAEE83 pKa = 4.45PGQGFNSLYY92 pKa = 10.17IPSARR97 pKa = 11.84DD98 pKa = 3.45YY99 pKa = 10.91YY100 pKa = 10.04NTNSSHH106 pKa = 6.11ARR108 pKa = 11.84SAEE111 pKa = 4.1TIFVRR116 pKa = 11.84GYY118 pKa = 9.64KK119 pKa = 9.88EE120 pKa = 3.87RR121 pKa = 11.84VNVTVSGGATWTWRR135 pKa = 11.84RR136 pKa = 11.84TVFCMKK142 pKa = 10.71GDD144 pKa = 3.86MLRR147 pKa = 11.84DD148 pKa = 3.11AWTDD152 pKa = 3.14NSQPPQYY159 pKa = 11.01DD160 pKa = 3.28SLGPMAGQTPTRR172 pKa = 11.84SIGPMDD178 pKa = 5.01TILADD183 pKa = 4.0VIRR186 pKa = 11.84GLLYY190 pKa = 10.63RR191 pKa = 11.84GVQGSDD197 pKa = 2.2WYY199 pKa = 10.59DD200 pKa = 3.81PYY202 pKa = 10.65TAPIDD207 pKa = 3.84TTRR210 pKa = 11.84VTLLRR215 pKa = 11.84DD216 pKa = 3.26QVITINPTNDD226 pKa = 2.86SGRR229 pKa = 11.84TRR231 pKa = 11.84MYY233 pKa = 10.75RR234 pKa = 11.84FWQPINKK241 pKa = 8.77NLAYY245 pKa = 10.03QSYY248 pKa = 9.78EE249 pKa = 3.85NDD251 pKa = 3.07EE252 pKa = 4.54STVSGAYY259 pKa = 8.88SVRR262 pKa = 11.84SKK264 pKa = 10.97PGIGDD269 pKa = 3.61IYY271 pKa = 10.72IFDD274 pKa = 3.56QVRR277 pKa = 11.84RR278 pKa = 11.84EE279 pKa = 4.02TGAATASFRR288 pKa = 11.84FSPEE292 pKa = 3.2GTFYY296 pKa = 9.87WHH298 pKa = 6.63EE299 pKa = 4.02RR300 pKa = 3.52
MM1 pKa = 7.66ALRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84LRR8 pKa = 11.84RR9 pKa = 11.84PTARR13 pKa = 11.84VRR15 pKa = 11.84PSRR18 pKa = 11.84ASGFRR23 pKa = 11.84RR24 pKa = 11.84VARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84FTRR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84IRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PMTNRR47 pKa = 11.84RR48 pKa = 11.84ILNVSSKK55 pKa = 10.86KK56 pKa = 10.48KK57 pKa = 9.95VDD59 pKa = 3.06TMMPLVIGNDD69 pKa = 3.85GVGTEE74 pKa = 4.49GPLLVTPAEE83 pKa = 4.45PGQGFNSLYY92 pKa = 10.17IPSARR97 pKa = 11.84DD98 pKa = 3.45YY99 pKa = 10.91YY100 pKa = 10.04NTNSSHH106 pKa = 6.11ARR108 pKa = 11.84SAEE111 pKa = 4.1TIFVRR116 pKa = 11.84GYY118 pKa = 9.64KK119 pKa = 9.88EE120 pKa = 3.87RR121 pKa = 11.84VNVTVSGGATWTWRR135 pKa = 11.84RR136 pKa = 11.84TVFCMKK142 pKa = 10.71GDD144 pKa = 3.86MLRR147 pKa = 11.84DD148 pKa = 3.11AWTDD152 pKa = 3.14NSQPPQYY159 pKa = 11.01DD160 pKa = 3.28SLGPMAGQTPTRR172 pKa = 11.84SIGPMDD178 pKa = 5.01TILADD183 pKa = 4.0VIRR186 pKa = 11.84GLLYY190 pKa = 10.63RR191 pKa = 11.84GVQGSDD197 pKa = 2.2WYY199 pKa = 10.59DD200 pKa = 3.81PYY202 pKa = 10.65TAPIDD207 pKa = 3.84TTRR210 pKa = 11.84VTLLRR215 pKa = 11.84DD216 pKa = 3.26QVITINPTNDD226 pKa = 2.86SGRR229 pKa = 11.84TRR231 pKa = 11.84MYY233 pKa = 10.75RR234 pKa = 11.84FWQPINKK241 pKa = 8.77NLAYY245 pKa = 10.03QSYY248 pKa = 9.78EE249 pKa = 3.85NDD251 pKa = 3.07EE252 pKa = 4.54STVSGAYY259 pKa = 8.88SVRR262 pKa = 11.84SKK264 pKa = 10.97PGIGDD269 pKa = 3.61IYY271 pKa = 10.72IFDD274 pKa = 3.56QVRR277 pKa = 11.84RR278 pKa = 11.84EE279 pKa = 4.02TGAATASFRR288 pKa = 11.84FSPEE292 pKa = 3.2GTFYY296 pKa = 9.87WHH298 pKa = 6.63EE299 pKa = 4.02RR300 pKa = 3.52
Molecular weight: 34.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
820 |
202 |
318 |
273.3 |
31.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.707 ± 0.316 | 1.951 ± 0.703 |
6.951 ± 0.639 | 5.732 ± 1.189 |
4.756 ± 0.625 | 7.805 ± 0.119 |
2.317 ± 0.716 | 5.122 ± 0.256 |
3.537 ± 0.607 | 6.463 ± 0.825 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.073 ± 0.402 | 2.927 ± 0.61 |
5.244 ± 0.498 | 2.561 ± 0.205 |
10.122 ± 1.716 | 5.366 ± 1.143 |
6.098 ± 1.293 | 7.073 ± 0.34 |
2.683 ± 0.303 | 4.512 ± 0.069 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
