
Farmington virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
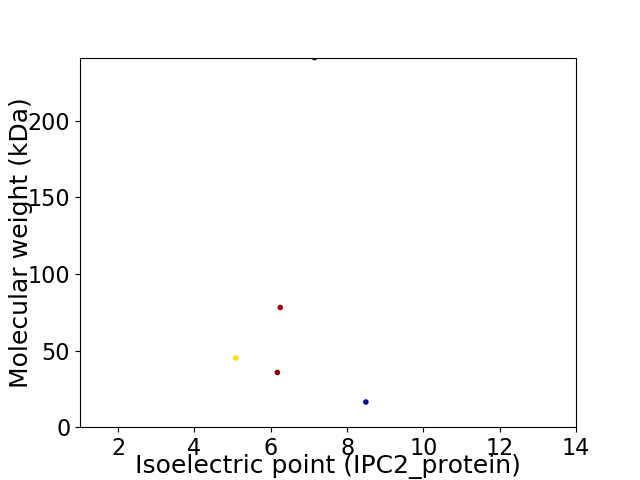
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|S4V3J8|S4V3J8_9RHAB G OS=Farmington virus OX=1027468 PE=4 SV=1
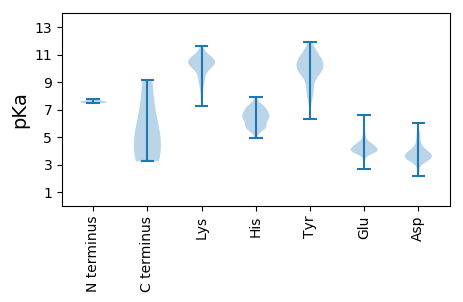
MM1 pKa = 7.54ARR3 pKa = 11.84PLAAAQHH10 pKa = 7.07LITEE14 pKa = 4.68RR15 pKa = 11.84HH16 pKa = 5.55SLQATLSRR24 pKa = 11.84ASKK27 pKa = 8.78TRR29 pKa = 11.84AEE31 pKa = 4.17EE32 pKa = 4.0FVKK35 pKa = 10.58DD36 pKa = 4.69FYY38 pKa = 11.5LQEE41 pKa = 4.18QYY43 pKa = 11.08SVPTIPTDD51 pKa = 5.29DD52 pKa = 3.74IAQSGPMLLQAILSEE67 pKa = 4.88EE68 pKa = 4.17YY69 pKa = 10.42TKK71 pKa = 10.65ATDD74 pKa = 3.2IAQSILWNTPTPNGLLRR91 pKa = 11.84EE92 pKa = 4.31HH93 pKa = 7.2LDD95 pKa = 3.42ADD97 pKa = 3.96GGGSFTALPASAIRR111 pKa = 11.84PSDD114 pKa = 3.61EE115 pKa = 3.75ANAWAARR122 pKa = 11.84ISDD125 pKa = 3.88SGLGPVFYY133 pKa = 10.3AALAAYY139 pKa = 9.47IIGWSGRR146 pKa = 11.84GEE148 pKa = 4.13TSRR151 pKa = 11.84VQQNIGQKK159 pKa = 9.17WLMNLNAIFGTTITHH174 pKa = 6.28PTTVRR179 pKa = 11.84LPINVVNNSLAVRR192 pKa = 11.84NGLAATLWLYY202 pKa = 10.6YY203 pKa = 10.01RR204 pKa = 11.84SSPQSQDD211 pKa = 2.35AFFYY215 pKa = 11.12GLIRR219 pKa = 11.84PCCSGYY225 pKa = 10.77LGLLHH230 pKa = 7.14RR231 pKa = 11.84VQEE234 pKa = 4.18IDD236 pKa = 3.34EE237 pKa = 4.46MEE239 pKa = 4.95PDD241 pKa = 3.94FLSDD245 pKa = 3.37PRR247 pKa = 11.84IIQVNEE253 pKa = 3.86VYY255 pKa = 10.43SALRR259 pKa = 11.84ALVQLGNDD267 pKa = 3.84FKK269 pKa = 10.86TADD272 pKa = 4.3DD273 pKa = 4.35EE274 pKa = 4.53PMQVWACRR282 pKa = 11.84GINNGYY288 pKa = 8.32LTYY291 pKa = 10.78LSEE294 pKa = 4.37TPAKK298 pKa = 9.89KK299 pKa = 10.3GAVVLMFAQCMLKK312 pKa = 10.4GDD314 pKa = 3.92SEE316 pKa = 4.44AWNSYY321 pKa = 7.6RR322 pKa = 11.84TATWVMPYY330 pKa = 9.85CDD332 pKa = 4.06NVALGAMAGYY342 pKa = 9.34IQARR346 pKa = 11.84QNTRR350 pKa = 11.84AYY352 pKa = 9.32EE353 pKa = 4.11VSAQTGLDD361 pKa = 3.36VNMAAVKK368 pKa = 10.53DD369 pKa = 4.85FEE371 pKa = 5.27ASSKK375 pKa = 10.49PKK377 pKa = 10.05AAPISLIPRR386 pKa = 11.84PADD389 pKa = 3.24VASRR393 pKa = 11.84TSEE396 pKa = 3.79RR397 pKa = 11.84PSIPEE402 pKa = 3.51VDD404 pKa = 3.48SDD406 pKa = 4.15EE407 pKa = 4.54EE408 pKa = 4.32LGGMM412 pKa = 4.8
MM1 pKa = 7.54ARR3 pKa = 11.84PLAAAQHH10 pKa = 7.07LITEE14 pKa = 4.68RR15 pKa = 11.84HH16 pKa = 5.55SLQATLSRR24 pKa = 11.84ASKK27 pKa = 8.78TRR29 pKa = 11.84AEE31 pKa = 4.17EE32 pKa = 4.0FVKK35 pKa = 10.58DD36 pKa = 4.69FYY38 pKa = 11.5LQEE41 pKa = 4.18QYY43 pKa = 11.08SVPTIPTDD51 pKa = 5.29DD52 pKa = 3.74IAQSGPMLLQAILSEE67 pKa = 4.88EE68 pKa = 4.17YY69 pKa = 10.42TKK71 pKa = 10.65ATDD74 pKa = 3.2IAQSILWNTPTPNGLLRR91 pKa = 11.84EE92 pKa = 4.31HH93 pKa = 7.2LDD95 pKa = 3.42ADD97 pKa = 3.96GGGSFTALPASAIRR111 pKa = 11.84PSDD114 pKa = 3.61EE115 pKa = 3.75ANAWAARR122 pKa = 11.84ISDD125 pKa = 3.88SGLGPVFYY133 pKa = 10.3AALAAYY139 pKa = 9.47IIGWSGRR146 pKa = 11.84GEE148 pKa = 4.13TSRR151 pKa = 11.84VQQNIGQKK159 pKa = 9.17WLMNLNAIFGTTITHH174 pKa = 6.28PTTVRR179 pKa = 11.84LPINVVNNSLAVRR192 pKa = 11.84NGLAATLWLYY202 pKa = 10.6YY203 pKa = 10.01RR204 pKa = 11.84SSPQSQDD211 pKa = 2.35AFFYY215 pKa = 11.12GLIRR219 pKa = 11.84PCCSGYY225 pKa = 10.77LGLLHH230 pKa = 7.14RR231 pKa = 11.84VQEE234 pKa = 4.18IDD236 pKa = 3.34EE237 pKa = 4.46MEE239 pKa = 4.95PDD241 pKa = 3.94FLSDD245 pKa = 3.37PRR247 pKa = 11.84IIQVNEE253 pKa = 3.86VYY255 pKa = 10.43SALRR259 pKa = 11.84ALVQLGNDD267 pKa = 3.84FKK269 pKa = 10.86TADD272 pKa = 4.3DD273 pKa = 4.35EE274 pKa = 4.53PMQVWACRR282 pKa = 11.84GINNGYY288 pKa = 8.32LTYY291 pKa = 10.78LSEE294 pKa = 4.37TPAKK298 pKa = 9.89KK299 pKa = 10.3GAVVLMFAQCMLKK312 pKa = 10.4GDD314 pKa = 3.92SEE316 pKa = 4.44AWNSYY321 pKa = 7.6RR322 pKa = 11.84TATWVMPYY330 pKa = 9.85CDD332 pKa = 4.06NVALGAMAGYY342 pKa = 9.34IQARR346 pKa = 11.84QNTRR350 pKa = 11.84AYY352 pKa = 9.32EE353 pKa = 4.11VSAQTGLDD361 pKa = 3.36VNMAAVKK368 pKa = 10.53DD369 pKa = 4.85FEE371 pKa = 5.27ASSKK375 pKa = 10.49PKK377 pKa = 10.05AAPISLIPRR386 pKa = 11.84PADD389 pKa = 3.24VASRR393 pKa = 11.84TSEE396 pKa = 3.79RR397 pKa = 11.84PSIPEE402 pKa = 3.51VDD404 pKa = 3.48SDD406 pKa = 4.15EE407 pKa = 4.54EE408 pKa = 4.32LGGMM412 pKa = 4.8
Molecular weight: 45.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4V2M6|S4V2M6_9RHAB Large structural protein OS=Farmington virus OX=1027468 PE=4 SV=1
MM1 pKa = 7.57RR2 pKa = 11.84RR3 pKa = 11.84FFLGEE8 pKa = 3.63SSAPARR14 pKa = 11.84DD15 pKa = 3.81WEE17 pKa = 4.57SEE19 pKa = 4.09RR20 pKa = 11.84PPPYY24 pKa = 10.15AVEE27 pKa = 4.49VPQSHH32 pKa = 7.24GIRR35 pKa = 11.84VTGYY39 pKa = 6.89FQCNEE44 pKa = 3.81RR45 pKa = 11.84PKK47 pKa = 10.76SKK49 pKa = 9.17KK50 pKa = 7.63TLHH53 pKa = 5.97SFAVKK58 pKa = 10.39LCDD61 pKa = 4.12AIKK64 pKa = 10.04PVRR67 pKa = 11.84ADD69 pKa = 3.4APSLKK74 pKa = 9.97IAIWTALDD82 pKa = 3.59LAFVKK87 pKa = 10.36PPNGTVTIDD96 pKa = 3.4AAVKK100 pKa = 8.37ATPLIGNTQYY110 pKa = 11.03TVGDD114 pKa = 4.64EE115 pKa = 3.95IFQMLGRR122 pKa = 11.84RR123 pKa = 11.84GGLIVIRR130 pKa = 11.84NLPHH134 pKa = 7.51DD135 pKa = 4.05YY136 pKa = 10.54PRR138 pKa = 11.84TLIEE142 pKa = 4.13FASPEE147 pKa = 4.0PP148 pKa = 3.81
MM1 pKa = 7.57RR2 pKa = 11.84RR3 pKa = 11.84FFLGEE8 pKa = 3.63SSAPARR14 pKa = 11.84DD15 pKa = 3.81WEE17 pKa = 4.57SEE19 pKa = 4.09RR20 pKa = 11.84PPPYY24 pKa = 10.15AVEE27 pKa = 4.49VPQSHH32 pKa = 7.24GIRR35 pKa = 11.84VTGYY39 pKa = 6.89FQCNEE44 pKa = 3.81RR45 pKa = 11.84PKK47 pKa = 10.76SKK49 pKa = 9.17KK50 pKa = 7.63TLHH53 pKa = 5.97SFAVKK58 pKa = 10.39LCDD61 pKa = 4.12AIKK64 pKa = 10.04PVRR67 pKa = 11.84ADD69 pKa = 3.4APSLKK74 pKa = 9.97IAIWTALDD82 pKa = 3.59LAFVKK87 pKa = 10.36PPNGTVTIDD96 pKa = 3.4AAVKK100 pKa = 8.37ATPLIGNTQYY110 pKa = 11.03TVGDD114 pKa = 4.64EE115 pKa = 3.95IFQMLGRR122 pKa = 11.84RR123 pKa = 11.84GGLIVIRR130 pKa = 11.84NLPHH134 pKa = 7.51DD135 pKa = 4.05YY136 pKa = 10.54PRR138 pKa = 11.84TLIEE142 pKa = 4.13FASPEE147 pKa = 4.0PP148 pKa = 3.81
Molecular weight: 16.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3709 |
148 |
2129 |
741.8 |
83.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.549 ± 1.317 | 1.645 ± 0.27 |
5.473 ± 0.154 | 5.985 ± 0.675 |
3.64 ± 0.418 | 5.5 ± 0.485 |
2.346 ± 0.357 | 5.985 ± 0.186 |
4.61 ± 0.709 | 10.299 ± 0.477 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.615 ± 0.358 | 3.586 ± 0.224 |
6.147 ± 0.423 | 2.858 ± 0.49 |
6.713 ± 0.24 | 7.522 ± 0.406 |
6.228 ± 0.064 | 6.471 ± 0.397 |
1.699 ± 0.097 | 3.128 ± 0.339 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
