
Bovine papillomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
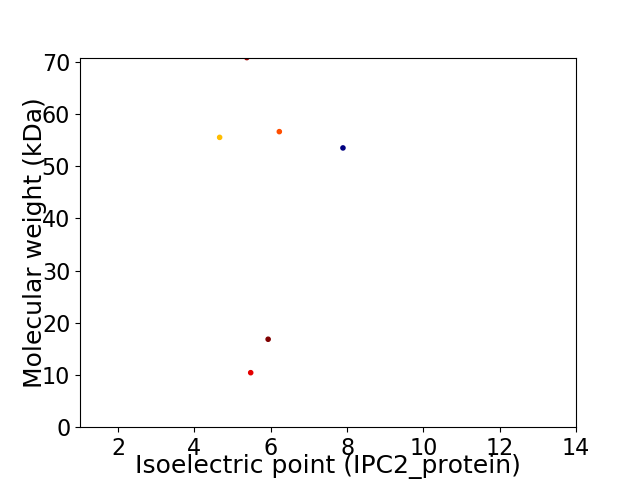
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z3FWK6|A0A1Z3FWK6_9PAPI Minor capsid protein L2 OS=Bovine papillomavirus OX=10571 GN=L2 PE=3 SV=1
MM1 pKa = 7.15SRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.78RR7 pKa = 11.84DD8 pKa = 3.18TVQNIYY14 pKa = 8.43NTCKK18 pKa = 10.05IWGNCPDD25 pKa = 3.9DD26 pKa = 4.54VINKK30 pKa = 9.25VEE32 pKa = 4.17SSTIADD38 pKa = 4.25KK39 pKa = 10.76ILKK42 pKa = 9.26YY43 pKa = 10.87GSGLVYY49 pKa = 10.45FGNAGIGTGSGGGGRR64 pKa = 11.84LGYY67 pKa = 10.4LPLGNRR73 pKa = 11.84PAGRR77 pKa = 11.84PAVPFSRR84 pKa = 11.84PTTRR88 pKa = 11.84VTTRR92 pKa = 11.84VPPIVEE98 pKa = 4.14TVVPDD103 pKa = 4.42SIAGNVVDD111 pKa = 5.16AGGPSVIDD119 pKa = 3.54LTDD122 pKa = 3.42LTTEE126 pKa = 3.96STGVDD131 pKa = 2.9VHH133 pKa = 6.89PDD135 pKa = 3.34VEE137 pKa = 4.62PGVLEE142 pKa = 4.68PGPTEE147 pKa = 4.07PPSIDD152 pKa = 3.44VTSDD156 pKa = 3.14TSPTIQVTEE165 pKa = 4.73SIHH168 pKa = 6.6SNPTFEE174 pKa = 4.17IPVVNGSTYY183 pKa = 8.57PTEE186 pKa = 4.27AVDD189 pKa = 3.47INTVSTVSTIIGTEE203 pKa = 3.92EE204 pKa = 3.92SAPLYY209 pKa = 10.25IPEE212 pKa = 4.56TIEE215 pKa = 3.83LQTFNTQRR223 pKa = 11.84TANNTFSEE231 pKa = 4.63TIIDD235 pKa = 3.66DD236 pKa = 3.81TGFDD240 pKa = 3.42SSTPLGKK247 pKa = 10.18RR248 pKa = 11.84PPEE251 pKa = 4.04GRR253 pKa = 11.84LGGARR258 pKa = 11.84VSVYY262 pKa = 10.32NKK264 pKa = 9.69RR265 pKa = 11.84YY266 pKa = 9.38AQVEE270 pKa = 4.32VEE272 pKa = 4.35NPAFIRR278 pKa = 11.84DD279 pKa = 4.12PYY281 pKa = 11.19TLVQYY286 pKa = 11.39SNVSYY291 pKa = 10.82DD292 pKa = 3.67PEE294 pKa = 4.15DD295 pKa = 3.35TVTFNRR301 pKa = 11.84GSGDD305 pKa = 3.42VNTAPDD311 pKa = 3.83PAFQDD316 pKa = 3.5VLKK319 pKa = 10.71LSKK322 pKa = 10.25PYY324 pKa = 7.6TTRR327 pKa = 11.84SPGGHH332 pKa = 5.2VRR334 pKa = 11.84LGRR337 pKa = 11.84VGQRR341 pKa = 11.84AGVKK345 pKa = 8.64TRR347 pKa = 11.84SGLQIGPQVHH357 pKa = 5.99YY358 pKa = 7.16FTRR361 pKa = 11.84ISSIDD366 pKa = 3.33PGEE369 pKa = 4.28TIEE372 pKa = 5.97LEE374 pKa = 4.34VLGDD378 pKa = 3.88PNTPDD383 pKa = 3.37VLVGDD388 pKa = 4.73DD389 pKa = 3.87EE390 pKa = 5.04FDD392 pKa = 3.29IVSLTEE398 pKa = 4.0SLADD402 pKa = 3.8SYY404 pKa = 11.87SEE406 pKa = 4.69SDD408 pKa = 3.74LLDD411 pKa = 3.45VLEE414 pKa = 5.87DD415 pKa = 3.12IGEE418 pKa = 4.08NLQLVMGGRR427 pKa = 11.84RR428 pKa = 11.84RR429 pKa = 11.84GPSVPVFTPRR439 pKa = 11.84GYY441 pKa = 10.07PPIEE445 pKa = 4.75SITVNFPGSGITIDD459 pKa = 3.69EE460 pKa = 4.36STAQSDD466 pKa = 4.01DD467 pKa = 3.76EE468 pKa = 4.8VPIEE472 pKa = 4.22PDD474 pKa = 3.03IPSTPSTPIRR484 pKa = 11.84PGLDD488 pKa = 3.07VYY490 pKa = 11.6DD491 pKa = 4.92SIDD494 pKa = 3.81FYY496 pKa = 11.47LHH498 pKa = 6.65PSLGKK503 pKa = 10.28KK504 pKa = 9.18LRR506 pKa = 11.84KK507 pKa = 8.27KK508 pKa = 10.1RR509 pKa = 11.84KK510 pKa = 7.99RR511 pKa = 11.84RR512 pKa = 11.84FYY514 pKa = 11.37
MM1 pKa = 7.15SRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.78RR7 pKa = 11.84DD8 pKa = 3.18TVQNIYY14 pKa = 8.43NTCKK18 pKa = 10.05IWGNCPDD25 pKa = 3.9DD26 pKa = 4.54VINKK30 pKa = 9.25VEE32 pKa = 4.17SSTIADD38 pKa = 4.25KK39 pKa = 10.76ILKK42 pKa = 9.26YY43 pKa = 10.87GSGLVYY49 pKa = 10.45FGNAGIGTGSGGGGRR64 pKa = 11.84LGYY67 pKa = 10.4LPLGNRR73 pKa = 11.84PAGRR77 pKa = 11.84PAVPFSRR84 pKa = 11.84PTTRR88 pKa = 11.84VTTRR92 pKa = 11.84VPPIVEE98 pKa = 4.14TVVPDD103 pKa = 4.42SIAGNVVDD111 pKa = 5.16AGGPSVIDD119 pKa = 3.54LTDD122 pKa = 3.42LTTEE126 pKa = 3.96STGVDD131 pKa = 2.9VHH133 pKa = 6.89PDD135 pKa = 3.34VEE137 pKa = 4.62PGVLEE142 pKa = 4.68PGPTEE147 pKa = 4.07PPSIDD152 pKa = 3.44VTSDD156 pKa = 3.14TSPTIQVTEE165 pKa = 4.73SIHH168 pKa = 6.6SNPTFEE174 pKa = 4.17IPVVNGSTYY183 pKa = 8.57PTEE186 pKa = 4.27AVDD189 pKa = 3.47INTVSTVSTIIGTEE203 pKa = 3.92EE204 pKa = 3.92SAPLYY209 pKa = 10.25IPEE212 pKa = 4.56TIEE215 pKa = 3.83LQTFNTQRR223 pKa = 11.84TANNTFSEE231 pKa = 4.63TIIDD235 pKa = 3.66DD236 pKa = 3.81TGFDD240 pKa = 3.42SSTPLGKK247 pKa = 10.18RR248 pKa = 11.84PPEE251 pKa = 4.04GRR253 pKa = 11.84LGGARR258 pKa = 11.84VSVYY262 pKa = 10.32NKK264 pKa = 9.69RR265 pKa = 11.84YY266 pKa = 9.38AQVEE270 pKa = 4.32VEE272 pKa = 4.35NPAFIRR278 pKa = 11.84DD279 pKa = 4.12PYY281 pKa = 11.19TLVQYY286 pKa = 11.39SNVSYY291 pKa = 10.82DD292 pKa = 3.67PEE294 pKa = 4.15DD295 pKa = 3.35TVTFNRR301 pKa = 11.84GSGDD305 pKa = 3.42VNTAPDD311 pKa = 3.83PAFQDD316 pKa = 3.5VLKK319 pKa = 10.71LSKK322 pKa = 10.25PYY324 pKa = 7.6TTRR327 pKa = 11.84SPGGHH332 pKa = 5.2VRR334 pKa = 11.84LGRR337 pKa = 11.84VGQRR341 pKa = 11.84AGVKK345 pKa = 8.64TRR347 pKa = 11.84SGLQIGPQVHH357 pKa = 5.99YY358 pKa = 7.16FTRR361 pKa = 11.84ISSIDD366 pKa = 3.33PGEE369 pKa = 4.28TIEE372 pKa = 5.97LEE374 pKa = 4.34VLGDD378 pKa = 3.88PNTPDD383 pKa = 3.37VLVGDD388 pKa = 4.73DD389 pKa = 3.87EE390 pKa = 5.04FDD392 pKa = 3.29IVSLTEE398 pKa = 4.0SLADD402 pKa = 3.8SYY404 pKa = 11.87SEE406 pKa = 4.69SDD408 pKa = 3.74LLDD411 pKa = 3.45VLEE414 pKa = 5.87DD415 pKa = 3.12IGEE418 pKa = 4.08NLQLVMGGRR427 pKa = 11.84RR428 pKa = 11.84RR429 pKa = 11.84GPSVPVFTPRR439 pKa = 11.84GYY441 pKa = 10.07PPIEE445 pKa = 4.75SITVNFPGSGITIDD459 pKa = 3.69EE460 pKa = 4.36STAQSDD466 pKa = 4.01DD467 pKa = 3.76EE468 pKa = 4.8VPIEE472 pKa = 4.22PDD474 pKa = 3.03IPSTPSTPIRR484 pKa = 11.84PGLDD488 pKa = 3.07VYY490 pKa = 11.6DD491 pKa = 4.92SIDD494 pKa = 3.81FYY496 pKa = 11.47LHH498 pKa = 6.65PSLGKK503 pKa = 10.28KK504 pKa = 9.18LRR506 pKa = 11.84KK507 pKa = 8.27KK508 pKa = 10.1RR509 pKa = 11.84KK510 pKa = 7.99RR511 pKa = 11.84RR512 pKa = 11.84FYY514 pKa = 11.37
Molecular weight: 55.54 kDa
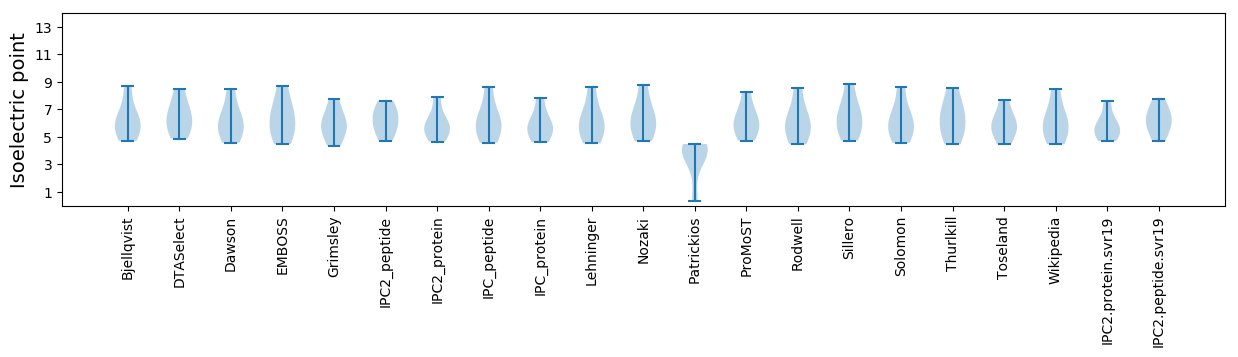
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z3FWF3|A0A1Z3FWF3_9PAPI Replication protein E1 OS=Bovine papillomavirus OX=10571 GN=E1 PE=3 SV=1
MM1 pKa = 7.8EE2 pKa = 5.38ILSARR7 pKa = 11.84LDD9 pKa = 3.71ALQEE13 pKa = 4.38DD14 pKa = 4.95LLSLYY19 pKa = 10.06EE20 pKa = 4.27SNKK23 pKa = 10.14NDD25 pKa = 2.98IDD27 pKa = 4.01SQIKK31 pKa = 9.43HH32 pKa = 5.03WMLLRR37 pKa = 11.84KK38 pKa = 9.6EE39 pKa = 4.22YY40 pKa = 10.49VIHH43 pKa = 6.43YY44 pKa = 6.79YY45 pKa = 10.95ARR47 pKa = 11.84ANSISRR53 pKa = 11.84LGYY56 pKa = 10.19QNILTQQVAQAKK68 pKa = 8.15AHH70 pKa = 5.74EE71 pKa = 4.93AIEE74 pKa = 4.16MVLNLEE80 pKa = 4.24TLKK83 pKa = 10.91ASPYY87 pKa = 10.42GAEE90 pKa = 3.88QWTLAQTSRR99 pKa = 11.84EE100 pKa = 3.98LWLTPPKK107 pKa = 10.58HH108 pKa = 6.02CFKK111 pKa = 11.04KK112 pKa = 10.29EE113 pKa = 3.66GSTVEE118 pKa = 3.64VWFDD122 pKa = 3.46GRR124 pKa = 11.84KK125 pKa = 9.93DD126 pKa = 3.51NAMHH130 pKa = 5.02YY131 pKa = 6.87TAWDD135 pKa = 3.65KK136 pKa = 10.91IYY138 pKa = 10.72YY139 pKa = 7.67QTDD142 pKa = 3.66SAWIKK147 pKa = 9.81TDD149 pKa = 2.81GHH151 pKa = 7.13VDD153 pKa = 3.38YY154 pKa = 10.84DD155 pKa = 3.65GCFYY159 pKa = 11.55YY160 pKa = 10.61EE161 pKa = 4.03GTYY164 pKa = 8.91KK165 pKa = 10.3HH166 pKa = 6.54YY167 pKa = 10.64YY168 pKa = 9.78IKK170 pKa = 10.61FQEE173 pKa = 4.65DD174 pKa = 3.18ADD176 pKa = 4.45KK177 pKa = 11.36YY178 pKa = 9.22SQSGDD183 pKa = 3.08WEE185 pKa = 4.5VHH187 pKa = 5.56FKK189 pKa = 10.66HH190 pKa = 6.74HH191 pKa = 6.08VFSPVASVSSTTDD204 pKa = 2.99GDD206 pKa = 4.11PDD208 pKa = 4.08PEE210 pKa = 5.86CGLRR214 pKa = 11.84ADD216 pKa = 3.72VQRR219 pKa = 11.84RR220 pKa = 11.84VPEE223 pKa = 3.91QHH225 pKa = 6.38QVPLDD230 pKa = 3.66NQATPRR236 pKa = 11.84KK237 pKa = 9.23RR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84DD241 pKa = 3.52EE242 pKa = 4.15TGGDD246 pKa = 3.42PAGPCTPRR254 pKa = 11.84RR255 pKa = 11.84KK256 pKa = 10.05ARR258 pKa = 11.84GDD260 pKa = 3.5RR261 pKa = 11.84CPHH264 pKa = 5.73SRR266 pKa = 11.84DD267 pKa = 3.06TRR269 pKa = 11.84GRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GRR275 pKa = 11.84DD276 pKa = 2.87RR277 pKa = 11.84GGGRR281 pKa = 11.84GGRR284 pKa = 11.84RR285 pKa = 11.84QRR287 pKa = 11.84RR288 pKa = 11.84EE289 pKa = 3.38GDD291 pKa = 3.43SEE293 pKa = 3.93EE294 pKa = 4.32RR295 pKa = 11.84QSQTEE300 pKa = 3.97DD301 pKa = 3.35SPDD304 pKa = 3.34EE305 pKa = 4.21RR306 pKa = 11.84FHH308 pKa = 7.1PGLTAAQQHH317 pKa = 5.63GQRR320 pKa = 11.84QRR322 pKa = 11.84TTAEE326 pKa = 4.16EE327 pKa = 4.68DD328 pKa = 3.37PHH330 pKa = 8.05SAAPLYY336 pKa = 10.23RR337 pKa = 11.84QEE339 pKa = 4.11EE340 pKa = 4.46GDD342 pKa = 3.4RR343 pKa = 11.84GEE345 pKa = 4.38ARR347 pKa = 11.84QPYY350 pKa = 8.57GSPRR354 pKa = 11.84IPEE357 pKa = 4.08EE358 pKa = 3.84PAQQIDD364 pKa = 4.13PNQLPRR370 pKa = 11.84VRR372 pKa = 11.84RR373 pKa = 11.84QPARR377 pKa = 11.84ILDD380 pKa = 3.91HH381 pKa = 7.06LADD384 pKa = 4.03TPILLLRR391 pKa = 11.84GGANQLKK398 pKa = 9.58CYY400 pKa = 10.27RR401 pKa = 11.84YY402 pKa = 9.85RR403 pKa = 11.84LLKK406 pKa = 10.81NYY408 pKa = 9.71FNLFLDD414 pKa = 5.17ISKK417 pKa = 7.8TWQWIGAGGIGNKK430 pKa = 9.79ARR432 pKa = 11.84VTVAFEE438 pKa = 4.18SLAQRR443 pKa = 11.84NQFLNTVPIPTGVTVAWGRR462 pKa = 11.84ISGLL466 pKa = 3.48
MM1 pKa = 7.8EE2 pKa = 5.38ILSARR7 pKa = 11.84LDD9 pKa = 3.71ALQEE13 pKa = 4.38DD14 pKa = 4.95LLSLYY19 pKa = 10.06EE20 pKa = 4.27SNKK23 pKa = 10.14NDD25 pKa = 2.98IDD27 pKa = 4.01SQIKK31 pKa = 9.43HH32 pKa = 5.03WMLLRR37 pKa = 11.84KK38 pKa = 9.6EE39 pKa = 4.22YY40 pKa = 10.49VIHH43 pKa = 6.43YY44 pKa = 6.79YY45 pKa = 10.95ARR47 pKa = 11.84ANSISRR53 pKa = 11.84LGYY56 pKa = 10.19QNILTQQVAQAKK68 pKa = 8.15AHH70 pKa = 5.74EE71 pKa = 4.93AIEE74 pKa = 4.16MVLNLEE80 pKa = 4.24TLKK83 pKa = 10.91ASPYY87 pKa = 10.42GAEE90 pKa = 3.88QWTLAQTSRR99 pKa = 11.84EE100 pKa = 3.98LWLTPPKK107 pKa = 10.58HH108 pKa = 6.02CFKK111 pKa = 11.04KK112 pKa = 10.29EE113 pKa = 3.66GSTVEE118 pKa = 3.64VWFDD122 pKa = 3.46GRR124 pKa = 11.84KK125 pKa = 9.93DD126 pKa = 3.51NAMHH130 pKa = 5.02YY131 pKa = 6.87TAWDD135 pKa = 3.65KK136 pKa = 10.91IYY138 pKa = 10.72YY139 pKa = 7.67QTDD142 pKa = 3.66SAWIKK147 pKa = 9.81TDD149 pKa = 2.81GHH151 pKa = 7.13VDD153 pKa = 3.38YY154 pKa = 10.84DD155 pKa = 3.65GCFYY159 pKa = 11.55YY160 pKa = 10.61EE161 pKa = 4.03GTYY164 pKa = 8.91KK165 pKa = 10.3HH166 pKa = 6.54YY167 pKa = 10.64YY168 pKa = 9.78IKK170 pKa = 10.61FQEE173 pKa = 4.65DD174 pKa = 3.18ADD176 pKa = 4.45KK177 pKa = 11.36YY178 pKa = 9.22SQSGDD183 pKa = 3.08WEE185 pKa = 4.5VHH187 pKa = 5.56FKK189 pKa = 10.66HH190 pKa = 6.74HH191 pKa = 6.08VFSPVASVSSTTDD204 pKa = 2.99GDD206 pKa = 4.11PDD208 pKa = 4.08PEE210 pKa = 5.86CGLRR214 pKa = 11.84ADD216 pKa = 3.72VQRR219 pKa = 11.84RR220 pKa = 11.84VPEE223 pKa = 3.91QHH225 pKa = 6.38QVPLDD230 pKa = 3.66NQATPRR236 pKa = 11.84KK237 pKa = 9.23RR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84DD241 pKa = 3.52EE242 pKa = 4.15TGGDD246 pKa = 3.42PAGPCTPRR254 pKa = 11.84RR255 pKa = 11.84KK256 pKa = 10.05ARR258 pKa = 11.84GDD260 pKa = 3.5RR261 pKa = 11.84CPHH264 pKa = 5.73SRR266 pKa = 11.84DD267 pKa = 3.06TRR269 pKa = 11.84GRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GRR275 pKa = 11.84DD276 pKa = 2.87RR277 pKa = 11.84GGGRR281 pKa = 11.84GGRR284 pKa = 11.84RR285 pKa = 11.84QRR287 pKa = 11.84RR288 pKa = 11.84EE289 pKa = 3.38GDD291 pKa = 3.43SEE293 pKa = 3.93EE294 pKa = 4.32RR295 pKa = 11.84QSQTEE300 pKa = 3.97DD301 pKa = 3.35SPDD304 pKa = 3.34EE305 pKa = 4.21RR306 pKa = 11.84FHH308 pKa = 7.1PGLTAAQQHH317 pKa = 5.63GQRR320 pKa = 11.84QRR322 pKa = 11.84TTAEE326 pKa = 4.16EE327 pKa = 4.68DD328 pKa = 3.37PHH330 pKa = 8.05SAAPLYY336 pKa = 10.23RR337 pKa = 11.84QEE339 pKa = 4.11EE340 pKa = 4.46GDD342 pKa = 3.4RR343 pKa = 11.84GEE345 pKa = 4.38ARR347 pKa = 11.84QPYY350 pKa = 8.57GSPRR354 pKa = 11.84IPEE357 pKa = 4.08EE358 pKa = 3.84PAQQIDD364 pKa = 4.13PNQLPRR370 pKa = 11.84VRR372 pKa = 11.84RR373 pKa = 11.84QPARR377 pKa = 11.84ILDD380 pKa = 3.91HH381 pKa = 7.06LADD384 pKa = 4.03TPILLLRR391 pKa = 11.84GGANQLKK398 pKa = 9.58CYY400 pKa = 10.27RR401 pKa = 11.84YY402 pKa = 9.85RR403 pKa = 11.84LLKK406 pKa = 10.81NYY408 pKa = 9.71FNLFLDD414 pKa = 5.17ISKK417 pKa = 7.8TWQWIGAGGIGNKK430 pKa = 9.79ARR432 pKa = 11.84VTVAFEE438 pKa = 4.18SLAQRR443 pKa = 11.84NQFLNTVPIPTGVTVAWGRR462 pKa = 11.84ISGLL466 pKa = 3.48
Molecular weight: 53.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2331 |
91 |
615 |
388.5 |
43.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.792 ± 0.73 | 2.402 ± 0.989 |
6.607 ± 0.309 | 6.349 ± 0.476 |
3.647 ± 0.436 | 6.65 ± 1.07 |
2.059 ± 0.582 | 4.59 ± 0.645 |
4.805 ± 0.57 | 8.237 ± 0.868 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.63 ± 0.545 | 4.505 ± 0.531 |
5.792 ± 1.078 | 5.234 ± 0.854 |
6.607 ± 0.82 | 6.735 ± 0.793 |
6.392 ± 1.064 | 6.392 ± 0.967 |
1.673 ± 0.461 | 3.904 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
