
Red clover powdery mildew-associated totivirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
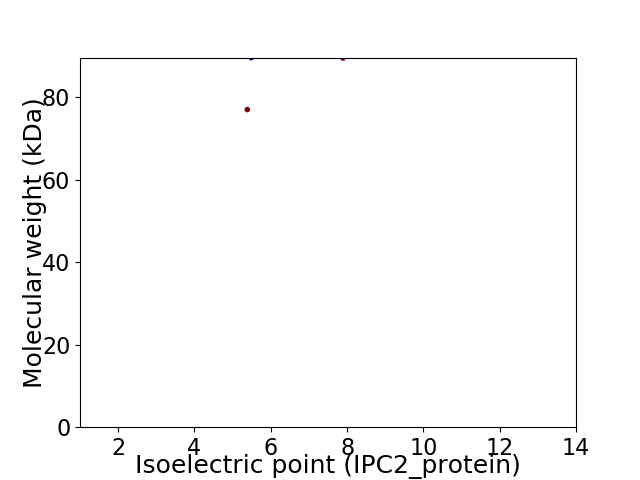
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S3Q2C1|A0A0S3Q2C1_9VIRU RNA-directed RNA polymerase OS=Red clover powdery mildew-associated totivirus 3 OX=1714364 GN=RdRp PE=3 SV=1
MM1 pKa = 7.29YY2 pKa = 10.25SQYY5 pKa = 11.13LDD7 pKa = 3.99TIFDD11 pKa = 3.59TSRR14 pKa = 11.84IANVVPYY21 pKa = 9.37VAKK24 pKa = 10.67GSYY27 pKa = 9.34TLVNKK32 pKa = 10.22ALAEE36 pKa = 4.16LKK38 pKa = 11.12VMDD41 pKa = 4.89RR42 pKa = 11.84SYY44 pKa = 11.51LSQLEE49 pKa = 4.22MSSDD53 pKa = 3.52FQVAVFEE60 pKa = 4.94PNVWTATASADD71 pKa = 3.36YY72 pKa = 10.81DD73 pKa = 4.0GYY75 pKa = 10.84NKK77 pKa = 10.15KK78 pKa = 10.35VLTSSGRR85 pKa = 11.84FSSSQAYY92 pKa = 9.94DD93 pKa = 3.22EE94 pKa = 4.43YY95 pKa = 11.38ARR97 pKa = 11.84SGSAIKK103 pKa = 9.91IQHH106 pKa = 6.65DD107 pKa = 3.26KK108 pKa = 10.8HH109 pKa = 7.86YY110 pKa = 10.89SIDD113 pKa = 3.53LTAGDD118 pKa = 4.72RR119 pKa = 11.84DD120 pKa = 3.85SQEE123 pKa = 4.22AFIFNMLVSWFKK135 pKa = 11.56AEE137 pKa = 3.85ITRR140 pKa = 11.84DD141 pKa = 3.93CEE143 pKa = 4.18PEE145 pKa = 3.89DD146 pKa = 3.69LFVRR150 pKa = 11.84QHH152 pKa = 5.41SFKK155 pKa = 10.75DD156 pKa = 3.51SHH158 pKa = 8.07VEE160 pKa = 3.44NAMADD165 pKa = 3.14GGMEE169 pKa = 3.98GKK171 pKa = 10.23VEE173 pKa = 3.88IRR175 pKa = 11.84LGAPAPEE182 pKa = 4.02RR183 pKa = 11.84LKK185 pKa = 10.91NALFVEE191 pKa = 4.49RR192 pKa = 11.84NDD194 pKa = 3.56LNFWTKK200 pKa = 10.46PYY202 pKa = 9.38VVKK205 pKa = 11.1YY206 pKa = 10.35NALTHH211 pKa = 5.63EE212 pKa = 4.07QQAFYY217 pKa = 9.91LAHH220 pKa = 5.74VMGRR224 pKa = 11.84TEE226 pKa = 4.17TDD228 pKa = 3.94GITADD233 pKa = 3.32ISIPGVMVQDD243 pKa = 4.45FLFEE247 pKa = 4.88PIGVEE252 pKa = 3.65LTRR255 pKa = 11.84DD256 pKa = 3.02IDD258 pKa = 3.88YY259 pKa = 11.49SSVDD263 pKa = 2.93WRR265 pKa = 11.84ASDD268 pKa = 4.89TMMAWIRR275 pKa = 11.84DD276 pKa = 3.75YY277 pKa = 11.86VEE279 pKa = 5.26LNRR282 pKa = 11.84CYY284 pKa = 10.52KK285 pKa = 10.64AFAAACDD292 pKa = 4.0LLGTLAFNPAPSFHH306 pKa = 6.94EE307 pKa = 4.65SIWWNPLVRR316 pKa = 11.84IVYY319 pKa = 7.81LAPFTPTRR327 pKa = 11.84ARR329 pKa = 11.84VPTTLNGEE337 pKa = 4.11GMYY340 pKa = 10.45EE341 pKa = 4.17SEE343 pKa = 4.42TILNFFRR350 pKa = 11.84NDD352 pKa = 2.99ACDD355 pKa = 3.29MKK357 pKa = 11.14NFIVMSSLVNYY368 pKa = 8.6ANHH371 pKa = 5.91MALYY375 pKa = 9.44GLLMNEE381 pKa = 4.37RR382 pKa = 11.84QDD384 pKa = 3.52LTDD387 pKa = 3.39WKK389 pKa = 9.86ICLSAIIGSMSNMRR403 pKa = 11.84GPLARR408 pKa = 11.84AQLFSLITNRR418 pKa = 11.84EE419 pKa = 4.11VEE421 pKa = 4.63SMTTNNCYY429 pKa = 10.5LEE431 pKa = 4.51YY432 pKa = 11.14DD433 pKa = 3.7MSDD436 pKa = 3.24MTGGALTVEE445 pKa = 4.45FDD447 pKa = 3.66VLKK450 pKa = 10.76GDD452 pKa = 3.92NQAPTAKK459 pKa = 9.79IINPVPYY466 pKa = 10.51VSGALFAGACGLDD479 pKa = 3.16VDD481 pKa = 4.48GFEE484 pKa = 6.11HH485 pKa = 7.06IMPRR489 pKa = 11.84QTIEE493 pKa = 3.69IPKK496 pKa = 10.16SGNLYY501 pKa = 9.88RR502 pKa = 11.84DD503 pKa = 3.34DD504 pKa = 4.1VFRR507 pKa = 11.84LAAAYY512 pKa = 9.83RR513 pKa = 11.84YY514 pKa = 9.16FGHH517 pKa = 6.62EE518 pKa = 4.07LDD520 pKa = 3.65ISAVRR525 pKa = 11.84DD526 pKa = 3.59RR527 pKa = 11.84TRR529 pKa = 11.84FSHH532 pKa = 5.61WCNGRR537 pKa = 11.84EE538 pKa = 3.95LVLAKK543 pKa = 10.66YY544 pKa = 9.45PILNAYY550 pKa = 10.19DD551 pKa = 4.16EE552 pKa = 4.23DD553 pKa = 4.29TKK555 pKa = 11.38FKK557 pKa = 10.71VHH559 pKa = 5.96GSKK562 pKa = 10.2RR563 pKa = 11.84RR564 pKa = 11.84PGRR567 pKa = 11.84HH568 pKa = 6.67DD569 pKa = 3.94DD570 pKa = 3.74VLEE573 pKa = 4.01ASYY576 pKa = 11.19LRR578 pKa = 11.84GGDD581 pKa = 3.49VVEE584 pKa = 4.65LVISRR589 pKa = 11.84PQIIMAGYY597 pKa = 6.68RR598 pKa = 11.84TRR600 pKa = 11.84RR601 pKa = 11.84EE602 pKa = 3.86IPTVASRR609 pKa = 11.84GVRR612 pKa = 11.84RR613 pKa = 11.84AKK615 pKa = 10.65VLLPKK620 pKa = 10.66VKK622 pKa = 10.27GAASSKK628 pKa = 9.23RR629 pKa = 11.84VSVRR633 pKa = 11.84ASALRR638 pKa = 11.84EE639 pKa = 3.73KK640 pKa = 10.75GDD642 pKa = 3.3RR643 pKa = 11.84HH644 pKa = 6.18EE645 pKa = 5.04SDD647 pKa = 3.62FPEE650 pKa = 4.51AQVGQAPSNPILNTSDD666 pKa = 4.03ASILEE671 pKa = 3.94QLNVDD676 pKa = 4.46LGQAAEE682 pKa = 4.15NAGG685 pKa = 3.51
MM1 pKa = 7.29YY2 pKa = 10.25SQYY5 pKa = 11.13LDD7 pKa = 3.99TIFDD11 pKa = 3.59TSRR14 pKa = 11.84IANVVPYY21 pKa = 9.37VAKK24 pKa = 10.67GSYY27 pKa = 9.34TLVNKK32 pKa = 10.22ALAEE36 pKa = 4.16LKK38 pKa = 11.12VMDD41 pKa = 4.89RR42 pKa = 11.84SYY44 pKa = 11.51LSQLEE49 pKa = 4.22MSSDD53 pKa = 3.52FQVAVFEE60 pKa = 4.94PNVWTATASADD71 pKa = 3.36YY72 pKa = 10.81DD73 pKa = 4.0GYY75 pKa = 10.84NKK77 pKa = 10.15KK78 pKa = 10.35VLTSSGRR85 pKa = 11.84FSSSQAYY92 pKa = 9.94DD93 pKa = 3.22EE94 pKa = 4.43YY95 pKa = 11.38ARR97 pKa = 11.84SGSAIKK103 pKa = 9.91IQHH106 pKa = 6.65DD107 pKa = 3.26KK108 pKa = 10.8HH109 pKa = 7.86YY110 pKa = 10.89SIDD113 pKa = 3.53LTAGDD118 pKa = 4.72RR119 pKa = 11.84DD120 pKa = 3.85SQEE123 pKa = 4.22AFIFNMLVSWFKK135 pKa = 11.56AEE137 pKa = 3.85ITRR140 pKa = 11.84DD141 pKa = 3.93CEE143 pKa = 4.18PEE145 pKa = 3.89DD146 pKa = 3.69LFVRR150 pKa = 11.84QHH152 pKa = 5.41SFKK155 pKa = 10.75DD156 pKa = 3.51SHH158 pKa = 8.07VEE160 pKa = 3.44NAMADD165 pKa = 3.14GGMEE169 pKa = 3.98GKK171 pKa = 10.23VEE173 pKa = 3.88IRR175 pKa = 11.84LGAPAPEE182 pKa = 4.02RR183 pKa = 11.84LKK185 pKa = 10.91NALFVEE191 pKa = 4.49RR192 pKa = 11.84NDD194 pKa = 3.56LNFWTKK200 pKa = 10.46PYY202 pKa = 9.38VVKK205 pKa = 11.1YY206 pKa = 10.35NALTHH211 pKa = 5.63EE212 pKa = 4.07QQAFYY217 pKa = 9.91LAHH220 pKa = 5.74VMGRR224 pKa = 11.84TEE226 pKa = 4.17TDD228 pKa = 3.94GITADD233 pKa = 3.32ISIPGVMVQDD243 pKa = 4.45FLFEE247 pKa = 4.88PIGVEE252 pKa = 3.65LTRR255 pKa = 11.84DD256 pKa = 3.02IDD258 pKa = 3.88YY259 pKa = 11.49SSVDD263 pKa = 2.93WRR265 pKa = 11.84ASDD268 pKa = 4.89TMMAWIRR275 pKa = 11.84DD276 pKa = 3.75YY277 pKa = 11.86VEE279 pKa = 5.26LNRR282 pKa = 11.84CYY284 pKa = 10.52KK285 pKa = 10.64AFAAACDD292 pKa = 4.0LLGTLAFNPAPSFHH306 pKa = 6.94EE307 pKa = 4.65SIWWNPLVRR316 pKa = 11.84IVYY319 pKa = 7.81LAPFTPTRR327 pKa = 11.84ARR329 pKa = 11.84VPTTLNGEE337 pKa = 4.11GMYY340 pKa = 10.45EE341 pKa = 4.17SEE343 pKa = 4.42TILNFFRR350 pKa = 11.84NDD352 pKa = 2.99ACDD355 pKa = 3.29MKK357 pKa = 11.14NFIVMSSLVNYY368 pKa = 8.6ANHH371 pKa = 5.91MALYY375 pKa = 9.44GLLMNEE381 pKa = 4.37RR382 pKa = 11.84QDD384 pKa = 3.52LTDD387 pKa = 3.39WKK389 pKa = 9.86ICLSAIIGSMSNMRR403 pKa = 11.84GPLARR408 pKa = 11.84AQLFSLITNRR418 pKa = 11.84EE419 pKa = 4.11VEE421 pKa = 4.63SMTTNNCYY429 pKa = 10.5LEE431 pKa = 4.51YY432 pKa = 11.14DD433 pKa = 3.7MSDD436 pKa = 3.24MTGGALTVEE445 pKa = 4.45FDD447 pKa = 3.66VLKK450 pKa = 10.76GDD452 pKa = 3.92NQAPTAKK459 pKa = 9.79IINPVPYY466 pKa = 10.51VSGALFAGACGLDD479 pKa = 3.16VDD481 pKa = 4.48GFEE484 pKa = 6.11HH485 pKa = 7.06IMPRR489 pKa = 11.84QTIEE493 pKa = 3.69IPKK496 pKa = 10.16SGNLYY501 pKa = 9.88RR502 pKa = 11.84DD503 pKa = 3.34DD504 pKa = 4.1VFRR507 pKa = 11.84LAAAYY512 pKa = 9.83RR513 pKa = 11.84YY514 pKa = 9.16FGHH517 pKa = 6.62EE518 pKa = 4.07LDD520 pKa = 3.65ISAVRR525 pKa = 11.84DD526 pKa = 3.59RR527 pKa = 11.84TRR529 pKa = 11.84FSHH532 pKa = 5.61WCNGRR537 pKa = 11.84EE538 pKa = 3.95LVLAKK543 pKa = 10.66YY544 pKa = 9.45PILNAYY550 pKa = 10.19DD551 pKa = 4.16EE552 pKa = 4.23DD553 pKa = 4.29TKK555 pKa = 11.38FKK557 pKa = 10.71VHH559 pKa = 5.96GSKK562 pKa = 10.2RR563 pKa = 11.84RR564 pKa = 11.84PGRR567 pKa = 11.84HH568 pKa = 6.67DD569 pKa = 3.94DD570 pKa = 3.74VLEE573 pKa = 4.01ASYY576 pKa = 11.19LRR578 pKa = 11.84GGDD581 pKa = 3.49VVEE584 pKa = 4.65LVISRR589 pKa = 11.84PQIIMAGYY597 pKa = 6.68RR598 pKa = 11.84TRR600 pKa = 11.84RR601 pKa = 11.84EE602 pKa = 3.86IPTVASRR609 pKa = 11.84GVRR612 pKa = 11.84RR613 pKa = 11.84AKK615 pKa = 10.65VLLPKK620 pKa = 10.66VKK622 pKa = 10.27GAASSKK628 pKa = 9.23RR629 pKa = 11.84VSVRR633 pKa = 11.84ASALRR638 pKa = 11.84EE639 pKa = 3.73KK640 pKa = 10.75GDD642 pKa = 3.3RR643 pKa = 11.84HH644 pKa = 6.18EE645 pKa = 5.04SDD647 pKa = 3.62FPEE650 pKa = 4.51AQVGQAPSNPILNTSDD666 pKa = 4.03ASILEE671 pKa = 3.94QLNVDD676 pKa = 4.46LGQAAEE682 pKa = 4.15NAGG685 pKa = 3.51
Molecular weight: 76.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3Q2C1|A0A0S3Q2C1_9VIRU RNA-directed RNA polymerase OS=Red clover powdery mildew-associated totivirus 3 OX=1714364 GN=RdRp PE=3 SV=1
MM1 pKa = 6.69TWKK4 pKa = 10.89AEE6 pKa = 4.36DD7 pKa = 4.15GVPMCIALAKK17 pKa = 10.71DD18 pKa = 3.4GANWNPSVFNLATHH32 pKa = 5.55VFARR36 pKa = 11.84VTEE39 pKa = 4.13SLAMEE44 pKa = 4.23GTEE47 pKa = 4.52VNTLLFGTITPAVHH61 pKa = 6.59IKK63 pKa = 10.34VGQLNCLYY71 pKa = 10.83LKK73 pKa = 10.06IDD75 pKa = 3.56QCCVTTHH82 pKa = 6.92PALNIILSRR91 pKa = 11.84HH92 pKa = 4.7YY93 pKa = 9.72SQLYY97 pKa = 8.96EE98 pKa = 4.18SVDD101 pKa = 4.04FVNPMSDD108 pKa = 2.5SMLFRR113 pKa = 11.84KK114 pKa = 10.04RR115 pKa = 11.84GINAEE120 pKa = 4.4DD121 pKa = 4.08GPEE124 pKa = 3.96PVRR127 pKa = 11.84GLLAMSKK134 pKa = 9.72LPHH137 pKa = 6.3SAITQQHH144 pKa = 6.19HH145 pKa = 4.98LHH147 pKa = 6.26FNITEE152 pKa = 4.26ACQTIDD158 pKa = 3.94RR159 pKa = 11.84NRR161 pKa = 11.84LSYY164 pKa = 10.76LLEE167 pKa = 4.25VLSVPSDD174 pKa = 3.26ATQAFASGLFMWGALVGDD192 pKa = 3.94AMFEE196 pKa = 4.13RR197 pKa = 11.84VRR199 pKa = 11.84EE200 pKa = 4.09SGLFGSRR207 pKa = 11.84SSAQFAAIAKK217 pKa = 8.72QISVEE222 pKa = 4.13AKK224 pKa = 9.98SLQNLHH230 pKa = 5.95TEE232 pKa = 4.47DD233 pKa = 3.48YY234 pKa = 10.95RR235 pKa = 11.84SIFEE239 pKa = 5.03LDD241 pKa = 3.35VLVNRR246 pKa = 11.84GVGKK250 pKa = 10.57VDD252 pKa = 3.23WEE254 pKa = 4.19KK255 pKa = 10.47EE256 pKa = 3.93QEE258 pKa = 4.07NRR260 pKa = 11.84QHH262 pKa = 6.55PKK264 pKa = 9.78VATVDD269 pKa = 3.14ACEE272 pKa = 4.16VRR274 pKa = 11.84RR275 pKa = 11.84TAYY278 pKa = 10.54LVFRR282 pKa = 11.84KK283 pKa = 9.96ALEE286 pKa = 3.79QGRR289 pKa = 11.84RR290 pKa = 11.84PVSMSWDD297 pKa = 3.96EE298 pKa = 3.86FWSARR303 pKa = 11.84WEE305 pKa = 3.91WTAAGAVHH313 pKa = 6.67SNAPGDD319 pKa = 3.52SKK321 pKa = 11.73YY322 pKa = 10.64IFKK325 pKa = 10.4EE326 pKa = 3.9VNEE329 pKa = 4.39MKK331 pKa = 10.82NKK333 pKa = 9.86FITMSAMPKK342 pKa = 10.22LPLSHH347 pKa = 7.5FLQRR351 pKa = 11.84EE352 pKa = 3.82PGIDD356 pKa = 2.73GWASVKK362 pKa = 10.48YY363 pKa = 9.61EE364 pKa = 3.78WGKK367 pKa = 9.96QRR369 pKa = 11.84AIYY372 pKa = 8.53GTDD375 pKa = 2.92LTSYY379 pKa = 10.02VLSNFAMYY387 pKa = 10.32ACEE390 pKa = 4.25SVLPSAFPVGPDD402 pKa = 3.01ADD404 pKa = 4.17EE405 pKa = 5.04ANVAARR411 pKa = 11.84VGGVLNNRR419 pKa = 11.84MPLCIDD425 pKa = 3.84FEE427 pKa = 4.92DD428 pKa = 5.15FNSQHH433 pKa = 6.14SVEE436 pKa = 4.58AMQAVLDD443 pKa = 4.57AYY445 pKa = 11.41SDD447 pKa = 3.98VFDD450 pKa = 4.61HH451 pKa = 7.13LLTVEE456 pKa = 3.95QKK458 pKa = 10.72YY459 pKa = 10.5ALRR462 pKa = 11.84WTAQSIGKK470 pKa = 8.21TSITNNIANCGTYY483 pKa = 7.94STKK486 pKa = 10.03GTMLSGWRR494 pKa = 11.84LTTMMNSVLNYY505 pKa = 10.19VYY507 pKa = 7.65TTHH510 pKa = 7.61LLGRR514 pKa = 11.84DD515 pKa = 3.39RR516 pKa = 11.84DD517 pKa = 4.2DD518 pKa = 3.63TPSVHH523 pKa = 6.78NGDD526 pKa = 4.41DD527 pKa = 3.38VLLGVKK533 pKa = 9.62NMRR536 pKa = 11.84VAQEE540 pKa = 3.88MNRR543 pKa = 11.84RR544 pKa = 11.84AQRR547 pKa = 11.84VGIRR551 pKa = 11.84LQPSKK556 pKa = 10.92CAFAGIAEE564 pKa = 4.62FLRR567 pKa = 11.84VDD569 pKa = 4.78RR570 pKa = 11.84IRR572 pKa = 11.84GSGGQYY578 pKa = 8.46LTRR581 pKa = 11.84SVATIVHH588 pKa = 5.87SRR590 pKa = 11.84VEE592 pKa = 4.58SKK594 pKa = 10.64PKK596 pKa = 10.47PNVKK600 pKa = 10.27DD601 pKa = 4.18LIEE604 pKa = 4.56AMEE607 pKa = 4.05TRR609 pKa = 11.84LADD612 pKa = 3.45LHH614 pKa = 6.37QRR616 pKa = 11.84GASIRR621 pKa = 11.84KK622 pKa = 7.2IARR625 pKa = 11.84IRR627 pKa = 11.84EE628 pKa = 4.14VYY630 pKa = 8.18YY631 pKa = 10.84HH632 pKa = 7.0KK633 pKa = 10.72LAGIFKK639 pKa = 10.25SEE641 pKa = 3.89VDD643 pKa = 2.95TMYY646 pKa = 10.73AIKK649 pKa = 10.36SIHH652 pKa = 6.22RR653 pKa = 11.84VNGGISQDD661 pKa = 2.9MYY663 pKa = 10.58AAVDD667 pKa = 3.75RR668 pKa = 11.84EE669 pKa = 4.18VLNVVNLEE677 pKa = 4.07KK678 pKa = 10.9SSVPEE683 pKa = 3.98LPKK686 pKa = 10.84LPGIHH691 pKa = 7.69DD692 pKa = 3.81YY693 pKa = 11.63ADD695 pKa = 3.55YY696 pKa = 10.18VAKK699 pKa = 10.48VLPIEE704 pKa = 4.4SKK706 pKa = 9.91VKK708 pKa = 10.42KK709 pKa = 9.41MCEE712 pKa = 3.87KK713 pKa = 9.7LTKK716 pKa = 9.38STQDD720 pKa = 3.18SVSMYY725 pKa = 8.52FTTAVLADD733 pKa = 3.86CTSVQDD739 pKa = 3.8RR740 pKa = 11.84LNMKK744 pKa = 10.08KK745 pKa = 9.83IYY747 pKa = 9.32KK748 pKa = 8.63AHH750 pKa = 6.43KK751 pKa = 10.28GEE753 pKa = 4.38IKK755 pKa = 10.22VSQYY759 pKa = 10.77GKK761 pKa = 10.7AVLTGYY767 pKa = 11.05GMEE770 pKa = 4.53LLRR773 pKa = 11.84SRR775 pKa = 11.84LRR777 pKa = 11.84PGALRR782 pKa = 11.84TLLTRR787 pKa = 11.84SEE789 pKa = 4.17NPIEE793 pKa = 4.15LLSLVVV799 pKa = 3.63
MM1 pKa = 6.69TWKK4 pKa = 10.89AEE6 pKa = 4.36DD7 pKa = 4.15GVPMCIALAKK17 pKa = 10.71DD18 pKa = 3.4GANWNPSVFNLATHH32 pKa = 5.55VFARR36 pKa = 11.84VTEE39 pKa = 4.13SLAMEE44 pKa = 4.23GTEE47 pKa = 4.52VNTLLFGTITPAVHH61 pKa = 6.59IKK63 pKa = 10.34VGQLNCLYY71 pKa = 10.83LKK73 pKa = 10.06IDD75 pKa = 3.56QCCVTTHH82 pKa = 6.92PALNIILSRR91 pKa = 11.84HH92 pKa = 4.7YY93 pKa = 9.72SQLYY97 pKa = 8.96EE98 pKa = 4.18SVDD101 pKa = 4.04FVNPMSDD108 pKa = 2.5SMLFRR113 pKa = 11.84KK114 pKa = 10.04RR115 pKa = 11.84GINAEE120 pKa = 4.4DD121 pKa = 4.08GPEE124 pKa = 3.96PVRR127 pKa = 11.84GLLAMSKK134 pKa = 9.72LPHH137 pKa = 6.3SAITQQHH144 pKa = 6.19HH145 pKa = 4.98LHH147 pKa = 6.26FNITEE152 pKa = 4.26ACQTIDD158 pKa = 3.94RR159 pKa = 11.84NRR161 pKa = 11.84LSYY164 pKa = 10.76LLEE167 pKa = 4.25VLSVPSDD174 pKa = 3.26ATQAFASGLFMWGALVGDD192 pKa = 3.94AMFEE196 pKa = 4.13RR197 pKa = 11.84VRR199 pKa = 11.84EE200 pKa = 4.09SGLFGSRR207 pKa = 11.84SSAQFAAIAKK217 pKa = 8.72QISVEE222 pKa = 4.13AKK224 pKa = 9.98SLQNLHH230 pKa = 5.95TEE232 pKa = 4.47DD233 pKa = 3.48YY234 pKa = 10.95RR235 pKa = 11.84SIFEE239 pKa = 5.03LDD241 pKa = 3.35VLVNRR246 pKa = 11.84GVGKK250 pKa = 10.57VDD252 pKa = 3.23WEE254 pKa = 4.19KK255 pKa = 10.47EE256 pKa = 3.93QEE258 pKa = 4.07NRR260 pKa = 11.84QHH262 pKa = 6.55PKK264 pKa = 9.78VATVDD269 pKa = 3.14ACEE272 pKa = 4.16VRR274 pKa = 11.84RR275 pKa = 11.84TAYY278 pKa = 10.54LVFRR282 pKa = 11.84KK283 pKa = 9.96ALEE286 pKa = 3.79QGRR289 pKa = 11.84RR290 pKa = 11.84PVSMSWDD297 pKa = 3.96EE298 pKa = 3.86FWSARR303 pKa = 11.84WEE305 pKa = 3.91WTAAGAVHH313 pKa = 6.67SNAPGDD319 pKa = 3.52SKK321 pKa = 11.73YY322 pKa = 10.64IFKK325 pKa = 10.4EE326 pKa = 3.9VNEE329 pKa = 4.39MKK331 pKa = 10.82NKK333 pKa = 9.86FITMSAMPKK342 pKa = 10.22LPLSHH347 pKa = 7.5FLQRR351 pKa = 11.84EE352 pKa = 3.82PGIDD356 pKa = 2.73GWASVKK362 pKa = 10.48YY363 pKa = 9.61EE364 pKa = 3.78WGKK367 pKa = 9.96QRR369 pKa = 11.84AIYY372 pKa = 8.53GTDD375 pKa = 2.92LTSYY379 pKa = 10.02VLSNFAMYY387 pKa = 10.32ACEE390 pKa = 4.25SVLPSAFPVGPDD402 pKa = 3.01ADD404 pKa = 4.17EE405 pKa = 5.04ANVAARR411 pKa = 11.84VGGVLNNRR419 pKa = 11.84MPLCIDD425 pKa = 3.84FEE427 pKa = 4.92DD428 pKa = 5.15FNSQHH433 pKa = 6.14SVEE436 pKa = 4.58AMQAVLDD443 pKa = 4.57AYY445 pKa = 11.41SDD447 pKa = 3.98VFDD450 pKa = 4.61HH451 pKa = 7.13LLTVEE456 pKa = 3.95QKK458 pKa = 10.72YY459 pKa = 10.5ALRR462 pKa = 11.84WTAQSIGKK470 pKa = 8.21TSITNNIANCGTYY483 pKa = 7.94STKK486 pKa = 10.03GTMLSGWRR494 pKa = 11.84LTTMMNSVLNYY505 pKa = 10.19VYY507 pKa = 7.65TTHH510 pKa = 7.61LLGRR514 pKa = 11.84DD515 pKa = 3.39RR516 pKa = 11.84DD517 pKa = 4.2DD518 pKa = 3.63TPSVHH523 pKa = 6.78NGDD526 pKa = 4.41DD527 pKa = 3.38VLLGVKK533 pKa = 9.62NMRR536 pKa = 11.84VAQEE540 pKa = 3.88MNRR543 pKa = 11.84RR544 pKa = 11.84AQRR547 pKa = 11.84VGIRR551 pKa = 11.84LQPSKK556 pKa = 10.92CAFAGIAEE564 pKa = 4.62FLRR567 pKa = 11.84VDD569 pKa = 4.78RR570 pKa = 11.84IRR572 pKa = 11.84GSGGQYY578 pKa = 8.46LTRR581 pKa = 11.84SVATIVHH588 pKa = 5.87SRR590 pKa = 11.84VEE592 pKa = 4.58SKK594 pKa = 10.64PKK596 pKa = 10.47PNVKK600 pKa = 10.27DD601 pKa = 4.18LIEE604 pKa = 4.56AMEE607 pKa = 4.05TRR609 pKa = 11.84LADD612 pKa = 3.45LHH614 pKa = 6.37QRR616 pKa = 11.84GASIRR621 pKa = 11.84KK622 pKa = 7.2IARR625 pKa = 11.84IRR627 pKa = 11.84EE628 pKa = 4.14VYY630 pKa = 8.18YY631 pKa = 10.84HH632 pKa = 7.0KK633 pKa = 10.72LAGIFKK639 pKa = 10.25SEE641 pKa = 3.89VDD643 pKa = 2.95TMYY646 pKa = 10.73AIKK649 pKa = 10.36SIHH652 pKa = 6.22RR653 pKa = 11.84VNGGISQDD661 pKa = 2.9MYY663 pKa = 10.58AAVDD667 pKa = 3.75RR668 pKa = 11.84EE669 pKa = 4.18VLNVVNLEE677 pKa = 4.07KK678 pKa = 10.9SSVPEE683 pKa = 3.98LPKK686 pKa = 10.84LPGIHH691 pKa = 7.69DD692 pKa = 3.81YY693 pKa = 11.63ADD695 pKa = 3.55YY696 pKa = 10.18VAKK699 pKa = 10.48VLPIEE704 pKa = 4.4SKK706 pKa = 9.91VKK708 pKa = 10.42KK709 pKa = 9.41MCEE712 pKa = 3.87KK713 pKa = 9.7LTKK716 pKa = 9.38STQDD720 pKa = 3.18SVSMYY725 pKa = 8.52FTTAVLADD733 pKa = 3.86CTSVQDD739 pKa = 3.8RR740 pKa = 11.84LNMKK744 pKa = 10.08KK745 pKa = 9.83IYY747 pKa = 9.32KK748 pKa = 8.63AHH750 pKa = 6.43KK751 pKa = 10.28GEE753 pKa = 4.38IKK755 pKa = 10.22VSQYY759 pKa = 10.77GKK761 pKa = 10.7AVLTGYY767 pKa = 11.05GMEE770 pKa = 4.53LLRR773 pKa = 11.84SRR775 pKa = 11.84LRR777 pKa = 11.84PGALRR782 pKa = 11.84TLLTRR787 pKa = 11.84SEE789 pKa = 4.17NPIEE793 pKa = 4.15LLSLVVV799 pKa = 3.63
Molecular weight: 89.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1484 |
685 |
799 |
742.0 |
83.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.03 ± 0.323 | 1.348 ± 0.126 |
5.93 ± 0.757 | 5.795 ± 0.031 |
3.774 ± 0.426 | 5.863 ± 0.016 |
2.426 ± 0.268 | 4.919 ± 0.134 |
4.919 ± 0.584 | 8.693 ± 0.363 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.302 ± 0.063 | 4.582 ± 0.165 |
3.841 ± 0.173 | 3.167 ± 0.276 |
6.469 ± 0.173 | 7.547 ± 0.072 |
5.256 ± 0.205 | 7.884 ± 0.513 |
1.415 ± 0.071 | 3.841 ± 0.378 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
