
Shewanella loihica (strain ATCC BAA-1088 / PV-4)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella; Shewanella loihica
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
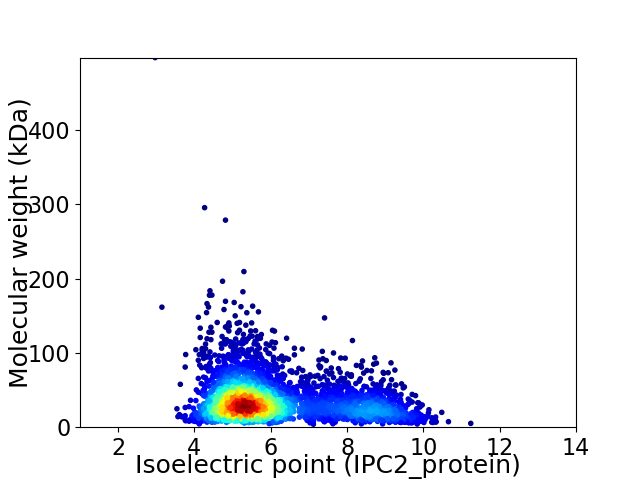
Virtual 2D-PAGE plot for 3855 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3QDI1|A3QDI1_SHELP NAD-dependent protein deacylase OS=Shewanella loihica (strain ATCC BAA-1088 / PV-4) OX=323850 GN=cobB PE=3 SV=1
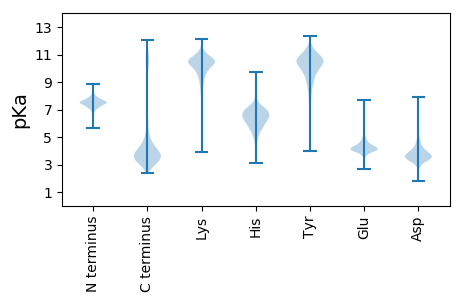
MM1 pKa = 7.66KK2 pKa = 10.35RR3 pKa = 11.84FKK5 pKa = 10.87CNRR8 pKa = 11.84LVRR11 pKa = 11.84ALGIVLISGASLYY24 pKa = 10.98GATITTANASFFPQKK39 pKa = 9.72MDD41 pKa = 3.53QACMAEE47 pKa = 3.91QAGFNLNCTANDD59 pKa = 3.38VRR61 pKa = 11.84VSKK64 pKa = 10.82VDD66 pKa = 3.92NIRR69 pKa = 11.84DD70 pKa = 3.88PNDD73 pKa = 3.3PNAKK77 pKa = 9.67VEE79 pKa = 4.38CNLGDD84 pKa = 3.88EE85 pKa = 4.65VTFIADD91 pKa = 3.36VTITTTANEE100 pKa = 4.21RR101 pKa = 11.84YY102 pKa = 9.41DD103 pKa = 3.72YY104 pKa = 11.16SVYY107 pKa = 10.55LPEE110 pKa = 5.19GNWSAQDD117 pKa = 3.51ANPNNEE123 pKa = 3.75CSILLGRR130 pKa = 11.84TDD132 pKa = 3.64GPGVDD137 pKa = 4.43LEE139 pKa = 4.7EE140 pKa = 5.52GLDD143 pKa = 3.49ACADD147 pKa = 3.22ISKK150 pKa = 10.48AAGYY154 pKa = 10.56NPTHH158 pKa = 6.66VYY160 pKa = 10.61ADD162 pKa = 3.55EE163 pKa = 5.13EE164 pKa = 4.19ITLFCRR170 pKa = 11.84DD171 pKa = 4.32DD172 pKa = 4.47DD173 pKa = 4.43NSGKK177 pKa = 10.63AEE179 pKa = 4.1FNYY182 pKa = 9.97CMAWHH187 pKa = 6.61NKK189 pKa = 7.86TGEE192 pKa = 4.36DD193 pKa = 4.59CSEE196 pKa = 5.07DD197 pKa = 4.09DD198 pKa = 3.92PAAPGTPSKK207 pKa = 10.59CRR209 pKa = 11.84CDD211 pKa = 3.75SFDD214 pKa = 3.34IDD216 pKa = 5.04VFIKK220 pKa = 10.54PNAPTITKK228 pKa = 9.79EE229 pKa = 4.26LIGTDD234 pKa = 3.1THH236 pKa = 6.18TEE238 pKa = 3.74PGGTYY243 pKa = 7.81TFKK246 pKa = 11.15LSFDD250 pKa = 3.72NPNAYY255 pKa = 8.9TSLFITDD262 pKa = 5.14LYY264 pKa = 11.6DD265 pKa = 4.43KK266 pKa = 10.79VDD268 pKa = 3.61EE269 pKa = 4.91GADD272 pKa = 3.52GSYY275 pKa = 9.25EE276 pKa = 4.15TMLDD280 pKa = 3.07LWNGVGAAGAADD292 pKa = 4.02GVYY295 pKa = 8.98LTASNCSQPANGGEE309 pKa = 4.12IVPSGSYY316 pKa = 9.99SCQFTVTIVDD326 pKa = 3.55RR327 pKa = 11.84DD328 pKa = 3.84LPDD331 pKa = 3.89DD332 pKa = 4.12QSPEE336 pKa = 4.09LYY338 pKa = 10.78NDD340 pKa = 3.95IIKK343 pKa = 10.59LALIDD348 pKa = 4.14KK349 pKa = 10.64NNDD352 pKa = 3.26PVVDD356 pKa = 4.31GGSCAAIGGVDD367 pKa = 4.19GDD369 pKa = 3.88HH370 pKa = 7.17CSNVLQVNVTNLPPSITVDD389 pKa = 3.18KK390 pKa = 11.02SAVPDD395 pKa = 3.54QVPEE399 pKa = 3.91SGAWVTYY406 pKa = 7.11TVRR409 pKa = 11.84VDD411 pKa = 3.35NTAADD416 pKa = 3.85YY417 pKa = 10.37DD418 pKa = 4.16SPLTLTYY425 pKa = 11.09LNDD428 pKa = 4.52DD429 pKa = 4.03KK430 pKa = 11.62FGDD433 pKa = 3.9LNGQGTCATGGTIAFGGYY451 pKa = 7.54YY452 pKa = 9.8EE453 pKa = 4.81CSFDD457 pKa = 4.62KK458 pKa = 10.93FISGTGAGSHH468 pKa = 6.16TNTATAKK475 pKa = 10.74AVDD478 pKa = 4.15DD479 pKa = 4.69EE480 pKa = 4.49NTEE483 pKa = 4.02AMNADD488 pKa = 3.69SATVIINDD496 pKa = 3.74IPSMITLEE504 pKa = 4.16KK505 pKa = 9.06TANPTSVLEE514 pKa = 4.39TGDD517 pKa = 3.54NPDD520 pKa = 3.1IYY522 pKa = 10.89RR523 pKa = 11.84DD524 pKa = 3.18VDD526 pKa = 3.87FTFWFSVDD534 pKa = 3.76DD535 pKa = 3.87QVGGQNTVDD544 pKa = 3.29SVTFATLTDD553 pKa = 4.94DD554 pKa = 3.48IFGVLTGDD562 pKa = 4.83CMVDD566 pKa = 3.31MKK568 pKa = 11.1NGAPIADD575 pKa = 3.79TPLAGFVLLPGEE587 pKa = 4.26NASCTITLQIQGYY600 pKa = 7.51RR601 pKa = 11.84TDD603 pKa = 2.69VHH605 pKa = 6.5TNVATIDD612 pKa = 3.72GTDD615 pKa = 3.37EE616 pKa = 4.77DD617 pKa = 4.48GQAVSDD623 pKa = 4.14SDD625 pKa = 3.93DD626 pKa = 3.44ATVTFTPAAPAVDD639 pKa = 3.71MDD641 pKa = 4.71FAATMLVVLGMHH653 pKa = 6.49NADD656 pKa = 3.49VNNANLTKK664 pKa = 9.92LTIGILDD671 pKa = 3.72VFAEE675 pKa = 4.37PDD677 pKa = 3.26TAGFKK682 pKa = 10.25IINGGGMYY690 pKa = 10.52NGTSYY695 pKa = 10.82GACGFNHH702 pKa = 6.88LFGYY706 pKa = 8.43TGSGTEE712 pKa = 4.4DD713 pKa = 3.71YY714 pKa = 10.35EE715 pKa = 4.54CAFTIEE721 pKa = 4.67LKK723 pKa = 10.68PGLEE727 pKa = 4.12NTDD730 pKa = 4.32PIAFLEE736 pKa = 4.26DD737 pKa = 3.37VVVEE741 pKa = 4.26LTNSQNDD748 pKa = 3.24KK749 pKa = 9.77STADD753 pKa = 3.22VSIQVGAIEE762 pKa = 3.89
MM1 pKa = 7.66KK2 pKa = 10.35RR3 pKa = 11.84FKK5 pKa = 10.87CNRR8 pKa = 11.84LVRR11 pKa = 11.84ALGIVLISGASLYY24 pKa = 10.98GATITTANASFFPQKK39 pKa = 9.72MDD41 pKa = 3.53QACMAEE47 pKa = 3.91QAGFNLNCTANDD59 pKa = 3.38VRR61 pKa = 11.84VSKK64 pKa = 10.82VDD66 pKa = 3.92NIRR69 pKa = 11.84DD70 pKa = 3.88PNDD73 pKa = 3.3PNAKK77 pKa = 9.67VEE79 pKa = 4.38CNLGDD84 pKa = 3.88EE85 pKa = 4.65VTFIADD91 pKa = 3.36VTITTTANEE100 pKa = 4.21RR101 pKa = 11.84YY102 pKa = 9.41DD103 pKa = 3.72YY104 pKa = 11.16SVYY107 pKa = 10.55LPEE110 pKa = 5.19GNWSAQDD117 pKa = 3.51ANPNNEE123 pKa = 3.75CSILLGRR130 pKa = 11.84TDD132 pKa = 3.64GPGVDD137 pKa = 4.43LEE139 pKa = 4.7EE140 pKa = 5.52GLDD143 pKa = 3.49ACADD147 pKa = 3.22ISKK150 pKa = 10.48AAGYY154 pKa = 10.56NPTHH158 pKa = 6.66VYY160 pKa = 10.61ADD162 pKa = 3.55EE163 pKa = 5.13EE164 pKa = 4.19ITLFCRR170 pKa = 11.84DD171 pKa = 4.32DD172 pKa = 4.47DD173 pKa = 4.43NSGKK177 pKa = 10.63AEE179 pKa = 4.1FNYY182 pKa = 9.97CMAWHH187 pKa = 6.61NKK189 pKa = 7.86TGEE192 pKa = 4.36DD193 pKa = 4.59CSEE196 pKa = 5.07DD197 pKa = 4.09DD198 pKa = 3.92PAAPGTPSKK207 pKa = 10.59CRR209 pKa = 11.84CDD211 pKa = 3.75SFDD214 pKa = 3.34IDD216 pKa = 5.04VFIKK220 pKa = 10.54PNAPTITKK228 pKa = 9.79EE229 pKa = 4.26LIGTDD234 pKa = 3.1THH236 pKa = 6.18TEE238 pKa = 3.74PGGTYY243 pKa = 7.81TFKK246 pKa = 11.15LSFDD250 pKa = 3.72NPNAYY255 pKa = 8.9TSLFITDD262 pKa = 5.14LYY264 pKa = 11.6DD265 pKa = 4.43KK266 pKa = 10.79VDD268 pKa = 3.61EE269 pKa = 4.91GADD272 pKa = 3.52GSYY275 pKa = 9.25EE276 pKa = 4.15TMLDD280 pKa = 3.07LWNGVGAAGAADD292 pKa = 4.02GVYY295 pKa = 8.98LTASNCSQPANGGEE309 pKa = 4.12IVPSGSYY316 pKa = 9.99SCQFTVTIVDD326 pKa = 3.55RR327 pKa = 11.84DD328 pKa = 3.84LPDD331 pKa = 3.89DD332 pKa = 4.12QSPEE336 pKa = 4.09LYY338 pKa = 10.78NDD340 pKa = 3.95IIKK343 pKa = 10.59LALIDD348 pKa = 4.14KK349 pKa = 10.64NNDD352 pKa = 3.26PVVDD356 pKa = 4.31GGSCAAIGGVDD367 pKa = 4.19GDD369 pKa = 3.88HH370 pKa = 7.17CSNVLQVNVTNLPPSITVDD389 pKa = 3.18KK390 pKa = 11.02SAVPDD395 pKa = 3.54QVPEE399 pKa = 3.91SGAWVTYY406 pKa = 7.11TVRR409 pKa = 11.84VDD411 pKa = 3.35NTAADD416 pKa = 3.85YY417 pKa = 10.37DD418 pKa = 4.16SPLTLTYY425 pKa = 11.09LNDD428 pKa = 4.52DD429 pKa = 4.03KK430 pKa = 11.62FGDD433 pKa = 3.9LNGQGTCATGGTIAFGGYY451 pKa = 7.54YY452 pKa = 9.8EE453 pKa = 4.81CSFDD457 pKa = 4.62KK458 pKa = 10.93FISGTGAGSHH468 pKa = 6.16TNTATAKK475 pKa = 10.74AVDD478 pKa = 4.15DD479 pKa = 4.69EE480 pKa = 4.49NTEE483 pKa = 4.02AMNADD488 pKa = 3.69SATVIINDD496 pKa = 3.74IPSMITLEE504 pKa = 4.16KK505 pKa = 9.06TANPTSVLEE514 pKa = 4.39TGDD517 pKa = 3.54NPDD520 pKa = 3.1IYY522 pKa = 10.89RR523 pKa = 11.84DD524 pKa = 3.18VDD526 pKa = 3.87FTFWFSVDD534 pKa = 3.76DD535 pKa = 3.87QVGGQNTVDD544 pKa = 3.29SVTFATLTDD553 pKa = 4.94DD554 pKa = 3.48IFGVLTGDD562 pKa = 4.83CMVDD566 pKa = 3.31MKK568 pKa = 11.1NGAPIADD575 pKa = 3.79TPLAGFVLLPGEE587 pKa = 4.26NASCTITLQIQGYY600 pKa = 7.51RR601 pKa = 11.84TDD603 pKa = 2.69VHH605 pKa = 6.5TNVATIDD612 pKa = 3.72GTDD615 pKa = 3.37EE616 pKa = 4.77DD617 pKa = 4.48GQAVSDD623 pKa = 4.14SDD625 pKa = 3.93DD626 pKa = 3.44ATVTFTPAAPAVDD639 pKa = 3.71MDD641 pKa = 4.71FAATMLVVLGMHH653 pKa = 6.49NADD656 pKa = 3.49VNNANLTKK664 pKa = 9.92LTIGILDD671 pKa = 3.72VFAEE675 pKa = 4.37PDD677 pKa = 3.26TAGFKK682 pKa = 10.25IINGGGMYY690 pKa = 10.52NGTSYY695 pKa = 10.82GACGFNHH702 pKa = 6.88LFGYY706 pKa = 8.43TGSGTEE712 pKa = 4.4DD713 pKa = 3.71YY714 pKa = 10.35EE715 pKa = 4.54CAFTIEE721 pKa = 4.67LKK723 pKa = 10.68PGLEE727 pKa = 4.12NTDD730 pKa = 4.32PIAFLEE736 pKa = 4.26DD737 pKa = 3.37VVVEE741 pKa = 4.26LTNSQNDD748 pKa = 3.24KK749 pKa = 9.77STADD753 pKa = 3.22VSIQVGAIEE762 pKa = 3.89
Molecular weight: 81.0 kDa
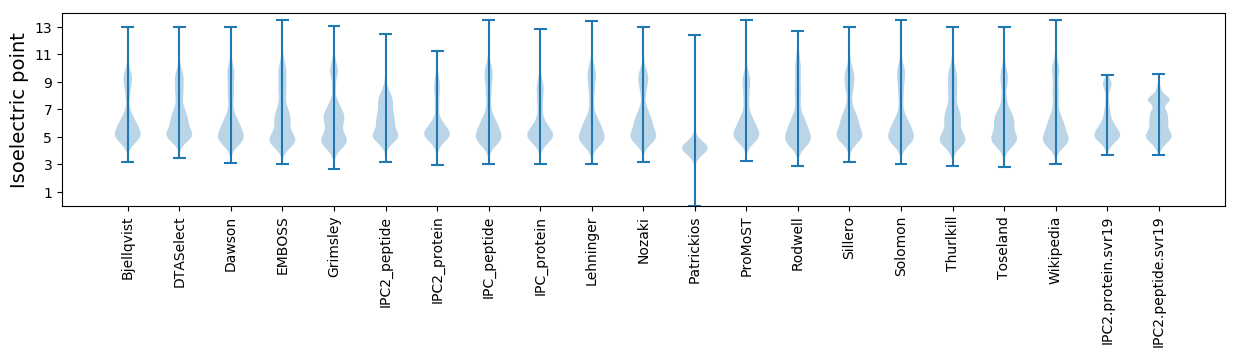
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3Q8W2|A3Q8W2_SHELP Phosphoesterase PA-phosphatase related OS=Shewanella loihica (strain ATCC BAA-1088 / PV-4) OX=323850 GN=Shew_0037 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84KK30 pKa = 9.3VIARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.35GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84KK30 pKa = 9.3VIARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.35GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1309847 |
34 |
4836 |
339.8 |
37.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.724 ± 0.045 | 1.051 ± 0.016 |
5.608 ± 0.039 | 6.111 ± 0.039 |
3.839 ± 0.026 | 7.273 ± 0.038 |
2.238 ± 0.023 | 5.759 ± 0.032 |
4.872 ± 0.032 | 10.984 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.626 ± 0.025 | 3.68 ± 0.036 |
4.086 ± 0.023 | 4.926 ± 0.039 |
4.775 ± 0.038 | 6.427 ± 0.039 |
5.041 ± 0.041 | 6.733 ± 0.038 |
1.212 ± 0.015 | 3.032 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
