
Acholeplasma laidlawii (strain PG-8A)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Acholeplasmatales; Acholeplasmataceae; Acholeplasma; Acholeplasma laidlawii
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
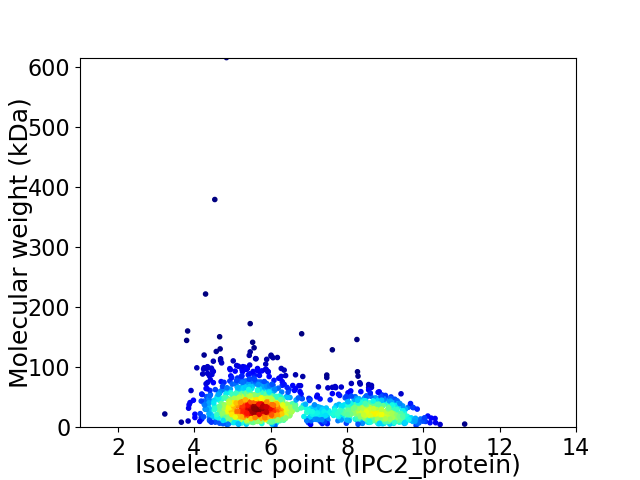
Virtual 2D-PAGE plot for 1380 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A9NHW7|A9NHW7_ACHLI [Ribosomal protein S18]-alanine N-acetyltransferase OS=Acholeplasma laidlawii (strain PG-8A) OX=441768 GN=rimI PE=3 SV=1
MM1 pKa = 7.47KK2 pKa = 9.9KK3 pKa = 9.95ISIIFTLLMTLVLIACTEE21 pKa = 3.85LTAYY25 pKa = 9.91KK26 pKa = 10.09IVFEE30 pKa = 4.59TNGGNDD36 pKa = 2.79IQAIEE41 pKa = 4.21FEE43 pKa = 4.34KK44 pKa = 10.72SYY46 pKa = 11.61ALAEE50 pKa = 4.11IEE52 pKa = 4.27KK53 pKa = 10.31VVPEE57 pKa = 3.91KK58 pKa = 10.72SGYY61 pKa = 7.79TFNSWYY67 pKa = 10.7LEE69 pKa = 3.79SDD71 pKa = 3.44LNEE74 pKa = 4.13GSKK77 pKa = 10.93LSTDD81 pKa = 2.88ITSSVTLYY89 pKa = 11.05AKK91 pKa = 8.33WTITEE96 pKa = 4.08YY97 pKa = 10.75TITLHH102 pKa = 7.04LDD104 pKa = 3.26GGVIHH109 pKa = 7.14PEE111 pKa = 3.86QVSKK115 pKa = 9.14FTILDD120 pKa = 3.73EE121 pKa = 4.66VNLLTPTKK129 pKa = 10.38EE130 pKa = 4.01GFKK133 pKa = 10.73FLGWSISNTEE143 pKa = 3.56ASFVDD148 pKa = 3.72KK149 pKa = 11.52VEE151 pKa = 4.3VGSTEE156 pKa = 3.96HH157 pKa = 6.74KK158 pKa = 10.47SFYY161 pKa = 11.1AKK163 pKa = 9.74WEE165 pKa = 3.91NLGEE169 pKa = 4.19VEE171 pKa = 4.13TFEE174 pKa = 5.96ISFKK178 pKa = 10.6NHH180 pKa = 6.49DD181 pKa = 4.0NAVLQTVEE189 pKa = 4.28VASGVVPAYY198 pKa = 9.42TGQTPTKK205 pKa = 9.49EE206 pKa = 3.75ATLTHH211 pKa = 5.91TFEE214 pKa = 4.04FTGWDD219 pKa = 3.5KK220 pKa = 11.59AVVAATADD228 pKa = 3.29IVYY231 pKa = 9.59VAQFKK236 pKa = 9.46EE237 pKa = 4.37VPITSGTTFNPALLNSIFGLDD258 pKa = 3.55VYY260 pKa = 11.52ALIPEE265 pKa = 4.69IVTSDD270 pKa = 3.75YY271 pKa = 11.51EE272 pKa = 4.73VIDD275 pKa = 3.8NSNDD279 pKa = 3.19LFNDD283 pKa = 3.63VYY285 pKa = 11.08IDD287 pKa = 3.61VFDD290 pKa = 3.72WTEE293 pKa = 4.09SDD295 pKa = 3.94LMAYY299 pKa = 9.56DD300 pKa = 4.38ALLEE304 pKa = 4.14NLLTYY309 pKa = 10.56DD310 pKa = 4.18ALEE313 pKa = 4.2DD314 pKa = 3.94AYY316 pKa = 10.07ILGEE320 pKa = 4.02LFIYY324 pKa = 10.08LYY326 pKa = 10.9ADD328 pKa = 3.84DD329 pKa = 5.66EE330 pKa = 4.56IVPGSIIYY338 pKa = 10.16GIGIYY343 pKa = 9.91QYY345 pKa = 11.3LEE347 pKa = 4.6DD348 pKa = 4.66EE349 pKa = 4.75TPVDD353 pKa = 3.53PVDD356 pKa = 3.58PVGAPFDD363 pKa = 4.0KK364 pKa = 11.19DD365 pKa = 3.39EE366 pKa = 5.08LNGIFGFDD374 pKa = 3.25IYY376 pKa = 11.55ALLPAIISEE385 pKa = 4.46DD386 pKa = 3.58VLITDD391 pKa = 5.33LSDD394 pKa = 2.97EE395 pKa = 4.67TYY397 pKa = 10.39IEE399 pKa = 4.35VYY401 pKa = 10.92VDD403 pKa = 3.17IFTWLDD409 pKa = 3.12ADD411 pKa = 3.85ADD413 pKa = 4.07AYY415 pKa = 10.84DD416 pKa = 4.14ALLSGSLAYY425 pKa = 10.26DD426 pKa = 3.55ATEE429 pKa = 4.08DD430 pKa = 3.6AYY432 pKa = 10.22KK433 pKa = 10.81LGDD436 pKa = 3.41YY437 pKa = 9.67FAYY440 pKa = 9.82IFIDD444 pKa = 3.51EE445 pKa = 4.22EE446 pKa = 4.91TYY448 pKa = 10.63PGLTVFGLAIYY459 pKa = 10.0GDD461 pKa = 3.92KK462 pKa = 11.24AGTTPVDD469 pKa = 3.29PVDD472 pKa = 3.62PVGAPFDD479 pKa = 4.0KK480 pKa = 11.19DD481 pKa = 3.39EE482 pKa = 5.08LNGIFGFDD490 pKa = 3.25IYY492 pKa = 11.51ALLPAIVSEE501 pKa = 4.61DD502 pKa = 3.57VLITDD507 pKa = 5.26LSDD510 pKa = 3.1EE511 pKa = 4.82TYY513 pKa = 10.73VEE515 pKa = 4.46VYY517 pKa = 11.01VDD519 pKa = 3.41IFTWLDD525 pKa = 3.12ADD527 pKa = 3.85ADD529 pKa = 4.07AYY531 pKa = 10.84DD532 pKa = 4.14ALLSGSLAYY541 pKa = 10.26DD542 pKa = 3.55ATEE545 pKa = 4.08DD546 pKa = 3.6AYY548 pKa = 10.22KK549 pKa = 10.81LGDD552 pKa = 3.41YY553 pKa = 9.67FAYY556 pKa = 9.82IFIDD560 pKa = 3.51EE561 pKa = 4.22EE562 pKa = 4.91TYY564 pKa = 10.63PGLTVFGLAIYY575 pKa = 10.0GDD577 pKa = 3.92KK578 pKa = 11.24AGTTPVDD585 pKa = 3.45PVEE588 pKa = 4.22PEE590 pKa = 3.18IGEE593 pKa = 4.27YY594 pKa = 11.06YY595 pKa = 10.59SFNVQDD601 pKa = 3.68TTSTLDD607 pKa = 3.05GSYY610 pKa = 10.73RR611 pKa = 11.84NNIDD615 pKa = 3.1VTLNFANNTNKK626 pKa = 10.01VIVKK630 pKa = 9.87ASHH633 pKa = 6.64IANITQTAPGGLSLGKK649 pKa = 10.0IFAANVSGNANPTVYY664 pKa = 10.91LEE666 pKa = 4.09IDD668 pKa = 3.2ALGNLIDD675 pKa = 3.84TMSFEE680 pKa = 4.19IQGRR684 pKa = 11.84TGFSPNLAGAKK695 pKa = 9.8LQVFNNGVWTDD706 pKa = 3.41LAGGNFYY713 pKa = 11.08SQIASSKK720 pKa = 8.41TLITISGINASKK732 pKa = 10.35FRR734 pKa = 11.84LLFQGTGATSNGGQFMIFNVNLLTGNAPAPVYY766 pKa = 9.29EE767 pKa = 4.3LWSDD771 pKa = 3.69VVTDD775 pKa = 4.94LEE777 pKa = 5.02AKK779 pKa = 10.42FDD781 pKa = 4.18DD782 pKa = 4.96LDD784 pKa = 4.3FNTYY788 pKa = 9.55MPDD791 pKa = 3.81FADD794 pKa = 3.65LTNLKK799 pKa = 7.99VTKK802 pKa = 10.68VSDD805 pKa = 3.22KK806 pKa = 10.31SFKK809 pKa = 10.77VVGSTTLDD817 pKa = 3.3VNTLYY822 pKa = 10.58TSYY825 pKa = 11.33INLILNKK832 pKa = 10.1SFEE835 pKa = 4.55KK836 pKa = 10.89NDD838 pKa = 3.65DD839 pKa = 3.51LSLVRR844 pKa = 11.84GHH846 pKa = 6.97DD847 pKa = 3.47VYY849 pKa = 11.73VYY851 pKa = 10.56VVNDD855 pKa = 3.73DD856 pKa = 3.84LAYY859 pKa = 10.74AMYY862 pKa = 9.92IIKK865 pKa = 8.86GTEE868 pKa = 3.74SLEE871 pKa = 3.99VYY873 pKa = 9.91IYY875 pKa = 10.83QFDD878 pKa = 3.7AVMDD882 pKa = 4.24DD883 pKa = 3.95VVLEE887 pKa = 4.28TLSKK891 pKa = 9.59RR892 pKa = 11.84QSINEE897 pKa = 4.0YY898 pKa = 9.69EE899 pKa = 4.21VSQFGMSGLPSTGTYY914 pKa = 10.42DD915 pKa = 3.51VLVVPVEE922 pKa = 4.0IQNVPFEE929 pKa = 4.43ASYY932 pKa = 8.51KK933 pKa = 9.13TKK935 pKa = 10.5LDD937 pKa = 3.28KK938 pKa = 11.34VFNGTSLDD946 pKa = 3.88TGWEE950 pKa = 4.49SVSSYY955 pKa = 10.64YY956 pKa = 10.64YY957 pKa = 10.36KK958 pKa = 10.85SSFGLLDD965 pKa = 4.27LNFDD969 pKa = 3.84ILDD972 pKa = 3.48KK973 pKa = 11.13HH974 pKa = 5.19VTSNVKK980 pKa = 10.27AFYY983 pKa = 9.81EE984 pKa = 4.6GKK986 pKa = 10.43GQDD989 pKa = 3.3GDD991 pKa = 3.54QYY993 pKa = 11.78AILDD997 pKa = 4.16ALTALDD1003 pKa = 3.71STIDD1007 pKa = 3.62FSKK1010 pKa = 11.1YY1011 pKa = 10.19DD1012 pKa = 3.71SNNDD1016 pKa = 3.22GVIDD1020 pKa = 4.37SIIFIYY1026 pKa = 9.26STDD1029 pKa = 3.24YY1030 pKa = 11.43NYY1032 pKa = 11.25DD1033 pKa = 3.36VDD1035 pKa = 4.85PWWAWVYY1042 pKa = 10.77VSNPDD1047 pKa = 3.0IVGSVSEE1054 pKa = 4.31LDD1056 pKa = 3.53GKK1058 pKa = 10.42NFEE1061 pKa = 4.51YY1062 pKa = 10.9YY1063 pKa = 10.13FWASYY1068 pKa = 11.68DD1069 pKa = 3.6FMNDD1073 pKa = 3.28ALPGNSDD1080 pKa = 4.96LILNSEE1086 pKa = 4.5TYY1088 pKa = 9.82IHH1090 pKa = 6.7EE1091 pKa = 5.01LGHH1094 pKa = 7.61LMGIVDD1100 pKa = 4.49FYY1102 pKa = 10.84PYY1104 pKa = 10.45EE1105 pKa = 4.38GNNQYY1110 pKa = 11.24GPMGGFDD1117 pKa = 3.37MMDD1120 pKa = 3.84YY1121 pKa = 10.91NAGDD1125 pKa = 3.51HH1126 pKa = 6.8GPFNKK1131 pKa = 10.17LVFGWLQPLVAQKK1144 pKa = 9.29GTYY1147 pKa = 9.1QVTLDD1152 pKa = 4.19SYY1154 pKa = 11.31STDD1157 pKa = 3.3TDD1159 pKa = 3.84GLNSTLLIPFNSSDD1173 pKa = 4.2LNDD1176 pKa = 3.32GNAFDD1181 pKa = 4.98EE1182 pKa = 4.47YY1183 pKa = 10.66LLVMFYY1189 pKa = 10.35TPNGLYY1195 pKa = 9.93SAHH1198 pKa = 6.86SGLEE1202 pKa = 4.11YY1203 pKa = 10.37IPSNAGIVVYY1213 pKa = 10.52HH1214 pKa = 6.56IDD1216 pKa = 4.49ARR1218 pKa = 11.84LTSNPVFWGEE1228 pKa = 3.67YY1229 pKa = 8.97FRR1231 pKa = 11.84NNNEE1235 pKa = 4.02GASSFINQILEE1246 pKa = 3.86ADD1248 pKa = 4.17KK1249 pKa = 11.52NNSIPGNGSIKK1260 pKa = 10.56QSDD1263 pKa = 4.4LLTSGTLNLNTYY1275 pKa = 8.9SWNQGGKK1282 pKa = 9.45INVSLTVASLFNNSSEE1298 pKa = 4.1NVTLTVSVAA1307 pKa = 3.15
MM1 pKa = 7.47KK2 pKa = 9.9KK3 pKa = 9.95ISIIFTLLMTLVLIACTEE21 pKa = 3.85LTAYY25 pKa = 9.91KK26 pKa = 10.09IVFEE30 pKa = 4.59TNGGNDD36 pKa = 2.79IQAIEE41 pKa = 4.21FEE43 pKa = 4.34KK44 pKa = 10.72SYY46 pKa = 11.61ALAEE50 pKa = 4.11IEE52 pKa = 4.27KK53 pKa = 10.31VVPEE57 pKa = 3.91KK58 pKa = 10.72SGYY61 pKa = 7.79TFNSWYY67 pKa = 10.7LEE69 pKa = 3.79SDD71 pKa = 3.44LNEE74 pKa = 4.13GSKK77 pKa = 10.93LSTDD81 pKa = 2.88ITSSVTLYY89 pKa = 11.05AKK91 pKa = 8.33WTITEE96 pKa = 4.08YY97 pKa = 10.75TITLHH102 pKa = 7.04LDD104 pKa = 3.26GGVIHH109 pKa = 7.14PEE111 pKa = 3.86QVSKK115 pKa = 9.14FTILDD120 pKa = 3.73EE121 pKa = 4.66VNLLTPTKK129 pKa = 10.38EE130 pKa = 4.01GFKK133 pKa = 10.73FLGWSISNTEE143 pKa = 3.56ASFVDD148 pKa = 3.72KK149 pKa = 11.52VEE151 pKa = 4.3VGSTEE156 pKa = 3.96HH157 pKa = 6.74KK158 pKa = 10.47SFYY161 pKa = 11.1AKK163 pKa = 9.74WEE165 pKa = 3.91NLGEE169 pKa = 4.19VEE171 pKa = 4.13TFEE174 pKa = 5.96ISFKK178 pKa = 10.6NHH180 pKa = 6.49DD181 pKa = 4.0NAVLQTVEE189 pKa = 4.28VASGVVPAYY198 pKa = 9.42TGQTPTKK205 pKa = 9.49EE206 pKa = 3.75ATLTHH211 pKa = 5.91TFEE214 pKa = 4.04FTGWDD219 pKa = 3.5KK220 pKa = 11.59AVVAATADD228 pKa = 3.29IVYY231 pKa = 9.59VAQFKK236 pKa = 9.46EE237 pKa = 4.37VPITSGTTFNPALLNSIFGLDD258 pKa = 3.55VYY260 pKa = 11.52ALIPEE265 pKa = 4.69IVTSDD270 pKa = 3.75YY271 pKa = 11.51EE272 pKa = 4.73VIDD275 pKa = 3.8NSNDD279 pKa = 3.19LFNDD283 pKa = 3.63VYY285 pKa = 11.08IDD287 pKa = 3.61VFDD290 pKa = 3.72WTEE293 pKa = 4.09SDD295 pKa = 3.94LMAYY299 pKa = 9.56DD300 pKa = 4.38ALLEE304 pKa = 4.14NLLTYY309 pKa = 10.56DD310 pKa = 4.18ALEE313 pKa = 4.2DD314 pKa = 3.94AYY316 pKa = 10.07ILGEE320 pKa = 4.02LFIYY324 pKa = 10.08LYY326 pKa = 10.9ADD328 pKa = 3.84DD329 pKa = 5.66EE330 pKa = 4.56IVPGSIIYY338 pKa = 10.16GIGIYY343 pKa = 9.91QYY345 pKa = 11.3LEE347 pKa = 4.6DD348 pKa = 4.66EE349 pKa = 4.75TPVDD353 pKa = 3.53PVDD356 pKa = 3.58PVGAPFDD363 pKa = 4.0KK364 pKa = 11.19DD365 pKa = 3.39EE366 pKa = 5.08LNGIFGFDD374 pKa = 3.25IYY376 pKa = 11.55ALLPAIISEE385 pKa = 4.46DD386 pKa = 3.58VLITDD391 pKa = 5.33LSDD394 pKa = 2.97EE395 pKa = 4.67TYY397 pKa = 10.39IEE399 pKa = 4.35VYY401 pKa = 10.92VDD403 pKa = 3.17IFTWLDD409 pKa = 3.12ADD411 pKa = 3.85ADD413 pKa = 4.07AYY415 pKa = 10.84DD416 pKa = 4.14ALLSGSLAYY425 pKa = 10.26DD426 pKa = 3.55ATEE429 pKa = 4.08DD430 pKa = 3.6AYY432 pKa = 10.22KK433 pKa = 10.81LGDD436 pKa = 3.41YY437 pKa = 9.67FAYY440 pKa = 9.82IFIDD444 pKa = 3.51EE445 pKa = 4.22EE446 pKa = 4.91TYY448 pKa = 10.63PGLTVFGLAIYY459 pKa = 10.0GDD461 pKa = 3.92KK462 pKa = 11.24AGTTPVDD469 pKa = 3.29PVDD472 pKa = 3.62PVGAPFDD479 pKa = 4.0KK480 pKa = 11.19DD481 pKa = 3.39EE482 pKa = 5.08LNGIFGFDD490 pKa = 3.25IYY492 pKa = 11.51ALLPAIVSEE501 pKa = 4.61DD502 pKa = 3.57VLITDD507 pKa = 5.26LSDD510 pKa = 3.1EE511 pKa = 4.82TYY513 pKa = 10.73VEE515 pKa = 4.46VYY517 pKa = 11.01VDD519 pKa = 3.41IFTWLDD525 pKa = 3.12ADD527 pKa = 3.85ADD529 pKa = 4.07AYY531 pKa = 10.84DD532 pKa = 4.14ALLSGSLAYY541 pKa = 10.26DD542 pKa = 3.55ATEE545 pKa = 4.08DD546 pKa = 3.6AYY548 pKa = 10.22KK549 pKa = 10.81LGDD552 pKa = 3.41YY553 pKa = 9.67FAYY556 pKa = 9.82IFIDD560 pKa = 3.51EE561 pKa = 4.22EE562 pKa = 4.91TYY564 pKa = 10.63PGLTVFGLAIYY575 pKa = 10.0GDD577 pKa = 3.92KK578 pKa = 11.24AGTTPVDD585 pKa = 3.45PVEE588 pKa = 4.22PEE590 pKa = 3.18IGEE593 pKa = 4.27YY594 pKa = 11.06YY595 pKa = 10.59SFNVQDD601 pKa = 3.68TTSTLDD607 pKa = 3.05GSYY610 pKa = 10.73RR611 pKa = 11.84NNIDD615 pKa = 3.1VTLNFANNTNKK626 pKa = 10.01VIVKK630 pKa = 9.87ASHH633 pKa = 6.64IANITQTAPGGLSLGKK649 pKa = 10.0IFAANVSGNANPTVYY664 pKa = 10.91LEE666 pKa = 4.09IDD668 pKa = 3.2ALGNLIDD675 pKa = 3.84TMSFEE680 pKa = 4.19IQGRR684 pKa = 11.84TGFSPNLAGAKK695 pKa = 9.8LQVFNNGVWTDD706 pKa = 3.41LAGGNFYY713 pKa = 11.08SQIASSKK720 pKa = 8.41TLITISGINASKK732 pKa = 10.35FRR734 pKa = 11.84LLFQGTGATSNGGQFMIFNVNLLTGNAPAPVYY766 pKa = 9.29EE767 pKa = 4.3LWSDD771 pKa = 3.69VVTDD775 pKa = 4.94LEE777 pKa = 5.02AKK779 pKa = 10.42FDD781 pKa = 4.18DD782 pKa = 4.96LDD784 pKa = 4.3FNTYY788 pKa = 9.55MPDD791 pKa = 3.81FADD794 pKa = 3.65LTNLKK799 pKa = 7.99VTKK802 pKa = 10.68VSDD805 pKa = 3.22KK806 pKa = 10.31SFKK809 pKa = 10.77VVGSTTLDD817 pKa = 3.3VNTLYY822 pKa = 10.58TSYY825 pKa = 11.33INLILNKK832 pKa = 10.1SFEE835 pKa = 4.55KK836 pKa = 10.89NDD838 pKa = 3.65DD839 pKa = 3.51LSLVRR844 pKa = 11.84GHH846 pKa = 6.97DD847 pKa = 3.47VYY849 pKa = 11.73VYY851 pKa = 10.56VVNDD855 pKa = 3.73DD856 pKa = 3.84LAYY859 pKa = 10.74AMYY862 pKa = 9.92IIKK865 pKa = 8.86GTEE868 pKa = 3.74SLEE871 pKa = 3.99VYY873 pKa = 9.91IYY875 pKa = 10.83QFDD878 pKa = 3.7AVMDD882 pKa = 4.24DD883 pKa = 3.95VVLEE887 pKa = 4.28TLSKK891 pKa = 9.59RR892 pKa = 11.84QSINEE897 pKa = 4.0YY898 pKa = 9.69EE899 pKa = 4.21VSQFGMSGLPSTGTYY914 pKa = 10.42DD915 pKa = 3.51VLVVPVEE922 pKa = 4.0IQNVPFEE929 pKa = 4.43ASYY932 pKa = 8.51KK933 pKa = 9.13TKK935 pKa = 10.5LDD937 pKa = 3.28KK938 pKa = 11.34VFNGTSLDD946 pKa = 3.88TGWEE950 pKa = 4.49SVSSYY955 pKa = 10.64YY956 pKa = 10.64YY957 pKa = 10.36KK958 pKa = 10.85SSFGLLDD965 pKa = 4.27LNFDD969 pKa = 3.84ILDD972 pKa = 3.48KK973 pKa = 11.13HH974 pKa = 5.19VTSNVKK980 pKa = 10.27AFYY983 pKa = 9.81EE984 pKa = 4.6GKK986 pKa = 10.43GQDD989 pKa = 3.3GDD991 pKa = 3.54QYY993 pKa = 11.78AILDD997 pKa = 4.16ALTALDD1003 pKa = 3.71STIDD1007 pKa = 3.62FSKK1010 pKa = 11.1YY1011 pKa = 10.19DD1012 pKa = 3.71SNNDD1016 pKa = 3.22GVIDD1020 pKa = 4.37SIIFIYY1026 pKa = 9.26STDD1029 pKa = 3.24YY1030 pKa = 11.43NYY1032 pKa = 11.25DD1033 pKa = 3.36VDD1035 pKa = 4.85PWWAWVYY1042 pKa = 10.77VSNPDD1047 pKa = 3.0IVGSVSEE1054 pKa = 4.31LDD1056 pKa = 3.53GKK1058 pKa = 10.42NFEE1061 pKa = 4.51YY1062 pKa = 10.9YY1063 pKa = 10.13FWASYY1068 pKa = 11.68DD1069 pKa = 3.6FMNDD1073 pKa = 3.28ALPGNSDD1080 pKa = 4.96LILNSEE1086 pKa = 4.5TYY1088 pKa = 9.82IHH1090 pKa = 6.7EE1091 pKa = 5.01LGHH1094 pKa = 7.61LMGIVDD1100 pKa = 4.49FYY1102 pKa = 10.84PYY1104 pKa = 10.45EE1105 pKa = 4.38GNNQYY1110 pKa = 11.24GPMGGFDD1117 pKa = 3.37MMDD1120 pKa = 3.84YY1121 pKa = 10.91NAGDD1125 pKa = 3.51HH1126 pKa = 6.8GPFNKK1131 pKa = 10.17LVFGWLQPLVAQKK1144 pKa = 9.29GTYY1147 pKa = 9.1QVTLDD1152 pKa = 4.19SYY1154 pKa = 11.31STDD1157 pKa = 3.3TDD1159 pKa = 3.84GLNSTLLIPFNSSDD1173 pKa = 4.2LNDD1176 pKa = 3.32GNAFDD1181 pKa = 4.98EE1182 pKa = 4.47YY1183 pKa = 10.66LLVMFYY1189 pKa = 10.35TPNGLYY1195 pKa = 9.93SAHH1198 pKa = 6.86SGLEE1202 pKa = 4.11YY1203 pKa = 10.37IPSNAGIVVYY1213 pKa = 10.52HH1214 pKa = 6.56IDD1216 pKa = 4.49ARR1218 pKa = 11.84LTSNPVFWGEE1228 pKa = 3.67YY1229 pKa = 8.97FRR1231 pKa = 11.84NNNEE1235 pKa = 4.02GASSFINQILEE1246 pKa = 3.86ADD1248 pKa = 4.17KK1249 pKa = 11.52NNSIPGNGSIKK1260 pKa = 10.56QSDD1263 pKa = 4.4LLTSGTLNLNTYY1275 pKa = 8.9SWNQGGKK1282 pKa = 9.45INVSLTVASLFNNSSEE1298 pKa = 4.1NVTLTVSVAA1307 pKa = 3.15
Molecular weight: 144.57 kDa
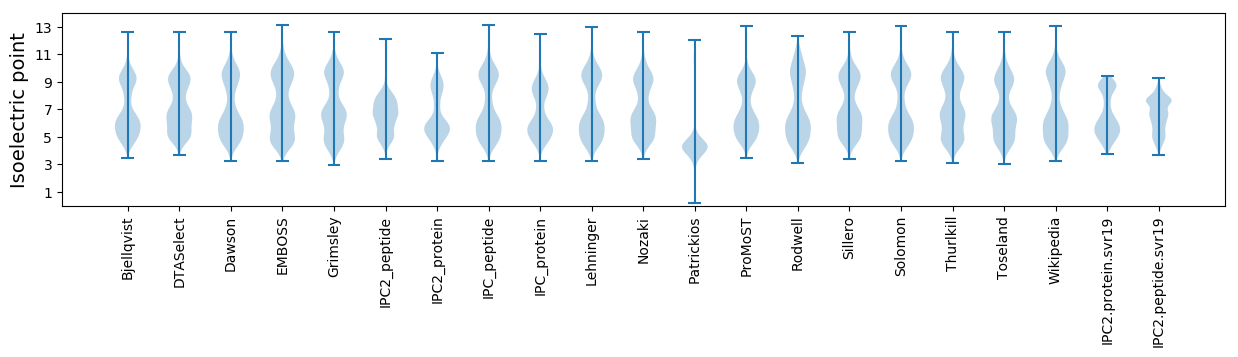
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9NE66|A9NE66_ACHLI IS-10 transposase OS=Acholeplasma laidlawii (strain PG-8A) OX=441768 GN=ACL_0003 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.95QPSKK9 pKa = 9.3IKK11 pKa = 9.77HH12 pKa = 4.29QRR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.25GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.24GRR39 pKa = 11.84QSLTVV44 pKa = 3.17
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.95QPSKK9 pKa = 9.3IKK11 pKa = 9.77HH12 pKa = 4.29QRR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.25GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.24GRR39 pKa = 11.84QSLTVV44 pKa = 3.17
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
451261 |
25 |
5552 |
327.0 |
37.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.75 ± 0.063 | 0.351 ± 0.012 |
6.011 ± 0.046 | 6.355 ± 0.05 |
4.843 ± 0.049 | 6.006 ± 0.079 |
2.089 ± 0.036 | 8.963 ± 0.064 |
7.82 ± 0.089 | 10.006 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.55 ± 0.031 | 5.534 ± 0.071 |
3.045 ± 0.032 | 3.106 ± 0.037 |
3.336 ± 0.048 | 6.139 ± 0.044 |
5.922 ± 0.075 | 6.846 ± 0.055 |
0.737 ± 0.023 | 4.591 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
