
Carrot yellow leaf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Closterovirus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
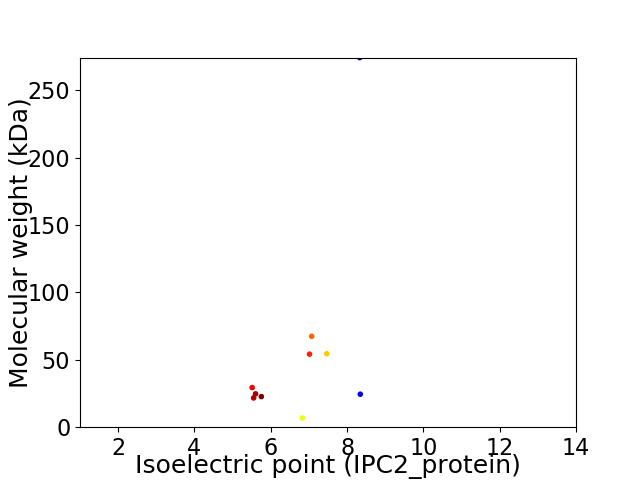
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
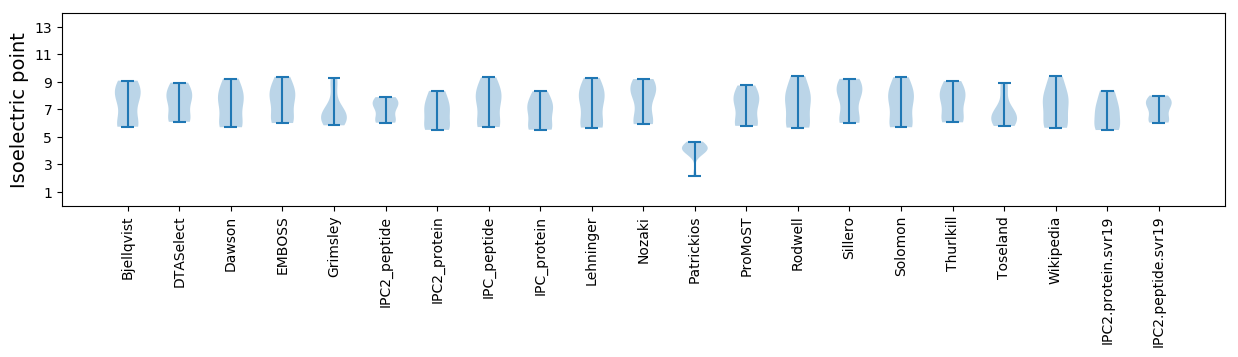
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GFK6|C6GFK6_9CLOS p25 OS=Carrot yellow leaf virus OX=656190 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.13LYY4 pKa = 10.36ISIYY8 pKa = 9.98EE9 pKa = 4.11YY10 pKa = 10.16IALVSEE16 pKa = 4.68ISRR19 pKa = 11.84LLDD22 pKa = 3.42LVSEE26 pKa = 4.64CVSGVTYY33 pKa = 10.83DD34 pKa = 2.99KK35 pKa = 11.01FKK37 pKa = 9.62EE38 pKa = 3.7FRR40 pKa = 11.84YY41 pKa = 10.13RR42 pKa = 11.84FVHH45 pKa = 5.87LTMLIPSLKK54 pKa = 10.22SDD56 pKa = 4.19LNDD59 pKa = 3.4SLRR62 pKa = 11.84DD63 pKa = 3.51EE64 pKa = 4.73GFSMQSEE71 pKa = 4.35KK72 pKa = 10.89KK73 pKa = 8.2EE74 pKa = 3.77QLVLFEE80 pKa = 4.69KK81 pKa = 10.08TCSDD85 pKa = 3.58VQSKK89 pKa = 8.99LRR91 pKa = 11.84RR92 pKa = 11.84MIVQSYY98 pKa = 10.01SISSPLDD105 pKa = 3.2LVVFFCKK112 pKa = 9.75KK113 pKa = 9.32HH114 pKa = 6.94YY115 pKa = 10.27EE116 pKa = 4.07MRR118 pKa = 11.84CCDD121 pKa = 3.99YY122 pKa = 10.74STIMHH127 pKa = 6.96DD128 pKa = 4.0KK129 pKa = 10.79VKK131 pKa = 10.1PSCDD135 pKa = 2.92AVLRR139 pKa = 11.84DD140 pKa = 3.3ISNTFGVDD148 pKa = 2.83VSPRR152 pKa = 11.84SFEE155 pKa = 3.95NGGFLTLKK163 pKa = 9.55DD164 pKa = 3.3TSFRR168 pKa = 11.84HH169 pKa = 5.71LFKK172 pKa = 11.05DD173 pKa = 3.54CFGVGEE179 pKa = 4.11EE180 pKa = 4.34EE181 pKa = 4.29IRR183 pKa = 11.84RR184 pKa = 11.84LLNEE188 pKa = 4.19RR189 pKa = 11.84EE190 pKa = 4.29TGSSLFSVEE199 pKa = 4.26PYY201 pKa = 8.7VPKK204 pKa = 10.63YY205 pKa = 10.02VSEE208 pKa = 4.21GAEE211 pKa = 3.85SHH213 pKa = 6.05II214 pKa = 4.85
MM1 pKa = 7.57KK2 pKa = 10.13LYY4 pKa = 10.36ISIYY8 pKa = 9.98EE9 pKa = 4.11YY10 pKa = 10.16IALVSEE16 pKa = 4.68ISRR19 pKa = 11.84LLDD22 pKa = 3.42LVSEE26 pKa = 4.64CVSGVTYY33 pKa = 10.83DD34 pKa = 2.99KK35 pKa = 11.01FKK37 pKa = 9.62EE38 pKa = 3.7FRR40 pKa = 11.84YY41 pKa = 10.13RR42 pKa = 11.84FVHH45 pKa = 5.87LTMLIPSLKK54 pKa = 10.22SDD56 pKa = 4.19LNDD59 pKa = 3.4SLRR62 pKa = 11.84DD63 pKa = 3.51EE64 pKa = 4.73GFSMQSEE71 pKa = 4.35KK72 pKa = 10.89KK73 pKa = 8.2EE74 pKa = 3.77QLVLFEE80 pKa = 4.69KK81 pKa = 10.08TCSDD85 pKa = 3.58VQSKK89 pKa = 8.99LRR91 pKa = 11.84RR92 pKa = 11.84MIVQSYY98 pKa = 10.01SISSPLDD105 pKa = 3.2LVVFFCKK112 pKa = 9.75KK113 pKa = 9.32HH114 pKa = 6.94YY115 pKa = 10.27EE116 pKa = 4.07MRR118 pKa = 11.84CCDD121 pKa = 3.99YY122 pKa = 10.74STIMHH127 pKa = 6.96DD128 pKa = 4.0KK129 pKa = 10.79VKK131 pKa = 10.1PSCDD135 pKa = 2.92AVLRR139 pKa = 11.84DD140 pKa = 3.3ISNTFGVDD148 pKa = 2.83VSPRR152 pKa = 11.84SFEE155 pKa = 3.95NGGFLTLKK163 pKa = 9.55DD164 pKa = 3.3TSFRR168 pKa = 11.84HH169 pKa = 5.71LFKK172 pKa = 11.05DD173 pKa = 3.54CFGVGEE179 pKa = 4.11EE180 pKa = 4.34EE181 pKa = 4.29IRR183 pKa = 11.84RR184 pKa = 11.84LLNEE188 pKa = 4.19RR189 pKa = 11.84EE190 pKa = 4.29TGSSLFSVEE199 pKa = 4.26PYY201 pKa = 8.7VPKK204 pKa = 10.63YY205 pKa = 10.02VSEE208 pKa = 4.21GAEE211 pKa = 3.85SHH213 pKa = 6.05II214 pKa = 4.85
Molecular weight: 24.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GFK4|C6GFK4_9CLOS p26 OS=Carrot yellow leaf virus OX=656190 PE=4 SV=1
MM1 pKa = 7.43SKK3 pKa = 10.61EE4 pKa = 3.93LMIVSPDD11 pKa = 3.3GSLEE15 pKa = 3.94KK16 pKa = 10.75ADD18 pKa = 3.7VSKK21 pKa = 10.96LAIATRR27 pKa = 11.84EE28 pKa = 4.0SMQAAFSALDD38 pKa = 3.75LSSVSGTSDD47 pKa = 2.98SCLSDD52 pKa = 4.03SEE54 pKa = 4.28LTEE57 pKa = 4.12AATKK61 pKa = 10.9VNTEE65 pKa = 4.09LKK67 pKa = 10.33KK68 pKa = 9.09ITKK71 pKa = 10.42GEE73 pKa = 4.54DD74 pKa = 2.72IDD76 pKa = 4.29MPSHH80 pKa = 6.24FAALIARR87 pKa = 11.84AATIGTSLSAVYY99 pKa = 10.22RR100 pKa = 11.84NQQTYY105 pKa = 10.18SIRR108 pKa = 11.84GKK110 pKa = 10.69GKK112 pKa = 8.46YY113 pKa = 6.27TVKK116 pKa = 10.45DD117 pKa = 3.59AEE119 pKa = 4.26IFPYY123 pKa = 10.35IIDD126 pKa = 3.36ITAKK130 pKa = 9.41YY131 pKa = 9.44GKK133 pKa = 9.42PNGLRR138 pKa = 11.84AFFASLEE145 pKa = 4.09NAFLVIAKK153 pKa = 7.54MKK155 pKa = 9.55PQLFEE160 pKa = 4.32SRR162 pKa = 11.84VATRR166 pKa = 11.84RR167 pKa = 11.84GTPKK171 pKa = 10.35EE172 pKa = 3.84KK173 pKa = 10.61GYY175 pKa = 10.52LATDD179 pKa = 4.53FLSGASPILVDD190 pKa = 4.09QEE192 pKa = 4.12RR193 pKa = 11.84AILNSASSYY202 pKa = 11.03ALDD205 pKa = 3.62RR206 pKa = 11.84AASKK210 pKa = 10.48KK211 pKa = 10.22KK212 pKa = 10.47NSGLVSLYY220 pKa = 10.77DD221 pKa = 3.46YY222 pKa = 11.41GRR224 pKa = 11.84YY225 pKa = 9.29DD226 pKa = 3.02
MM1 pKa = 7.43SKK3 pKa = 10.61EE4 pKa = 3.93LMIVSPDD11 pKa = 3.3GSLEE15 pKa = 3.94KK16 pKa = 10.75ADD18 pKa = 3.7VSKK21 pKa = 10.96LAIATRR27 pKa = 11.84EE28 pKa = 4.0SMQAAFSALDD38 pKa = 3.75LSSVSGTSDD47 pKa = 2.98SCLSDD52 pKa = 4.03SEE54 pKa = 4.28LTEE57 pKa = 4.12AATKK61 pKa = 10.9VNTEE65 pKa = 4.09LKK67 pKa = 10.33KK68 pKa = 9.09ITKK71 pKa = 10.42GEE73 pKa = 4.54DD74 pKa = 2.72IDD76 pKa = 4.29MPSHH80 pKa = 6.24FAALIARR87 pKa = 11.84AATIGTSLSAVYY99 pKa = 10.22RR100 pKa = 11.84NQQTYY105 pKa = 10.18SIRR108 pKa = 11.84GKK110 pKa = 10.69GKK112 pKa = 8.46YY113 pKa = 6.27TVKK116 pKa = 10.45DD117 pKa = 3.59AEE119 pKa = 4.26IFPYY123 pKa = 10.35IIDD126 pKa = 3.36ITAKK130 pKa = 9.41YY131 pKa = 9.44GKK133 pKa = 9.42PNGLRR138 pKa = 11.84AFFASLEE145 pKa = 4.09NAFLVIAKK153 pKa = 7.54MKK155 pKa = 9.55PQLFEE160 pKa = 4.32SRR162 pKa = 11.84VATRR166 pKa = 11.84RR167 pKa = 11.84GTPKK171 pKa = 10.35EE172 pKa = 3.84KK173 pKa = 10.61GYY175 pKa = 10.52LATDD179 pKa = 4.53FLSGASPILVDD190 pKa = 4.09QEE192 pKa = 4.12RR193 pKa = 11.84AILNSASSYY202 pKa = 11.03ALDD205 pKa = 3.62RR206 pKa = 11.84AASKK210 pKa = 10.48KK211 pKa = 10.22KK212 pKa = 10.47NSGLVSLYY220 pKa = 10.77DD221 pKa = 3.46YY222 pKa = 11.41GRR224 pKa = 11.84YY225 pKa = 9.29DD226 pKa = 3.02
Molecular weight: 24.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5163 |
60 |
2431 |
516.3 |
58.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.043 ± 0.568 | 2.247 ± 0.183 |
5.404 ± 0.175 | 5.559 ± 0.261 |
5.907 ± 0.436 | 5.075 ± 0.408 |
2.305 ± 0.273 | 5.539 ± 0.32 |
6.488 ± 0.322 | 9.607 ± 0.262 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.146 | 3.68 ± 0.249 |
4.009 ± 0.247 | 2.382 ± 0.334 |
5.559 ± 0.555 | 11.059 ± 0.318 |
5.617 ± 0.325 | 7.011 ± 0.376 |
0.542 ± 0.179 | 4.048 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
