
Cumuto virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Goukovirus; Cumuto goukovirus
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
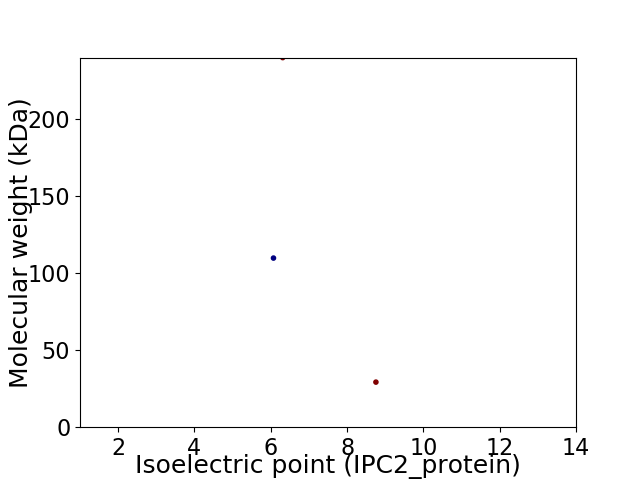
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|W5VH60|W5VH60_9VIRU Glycoprotein G OS=Cumuto virus OX=1457166 PE=4 SV=1
MM1 pKa = 7.68ASVKK5 pKa = 10.42LLFVIYY11 pKa = 9.81CISSLMINEE20 pKa = 3.94MRR22 pKa = 11.84TVEE25 pKa = 5.16LPEE28 pKa = 3.97FWHH31 pKa = 6.3MSRR34 pKa = 11.84EE35 pKa = 3.94KK36 pKa = 10.35MNRR39 pKa = 11.84KK40 pKa = 9.26PIDD43 pKa = 3.64PNVKK47 pKa = 10.01VKK49 pKa = 10.79GGNCSNGVPANATIQDD65 pKa = 3.61EE66 pKa = 5.17GITLNKK72 pKa = 9.82CSKK75 pKa = 10.75QMTAVVRR82 pKa = 11.84YY83 pKa = 6.61TVEE86 pKa = 4.23GVDD89 pKa = 3.36EE90 pKa = 4.41TFGCVVNYY98 pKa = 10.61DD99 pKa = 3.88CGLNLHH105 pKa = 6.63NGDD108 pKa = 4.82GYY110 pKa = 10.23CTLKK114 pKa = 11.15NEE116 pKa = 4.62DD117 pKa = 4.34CFHH120 pKa = 6.88EE121 pKa = 4.81PGCIVPKK128 pKa = 10.7DD129 pKa = 4.06RR130 pKa = 11.84ILPMGNCNEE139 pKa = 4.34THH141 pKa = 5.39WTQNRR146 pKa = 11.84PEE148 pKa = 4.06FCSVGVHH155 pKa = 6.38KK156 pKa = 10.72AGCEE160 pKa = 3.62EE161 pKa = 3.97TGRR164 pKa = 11.84VVRR167 pKa = 11.84LPWVVDD173 pKa = 3.72RR174 pKa = 11.84SGKK177 pKa = 9.74KK178 pKa = 9.96RR179 pKa = 11.84LINKK183 pKa = 9.23LNIEE187 pKa = 4.39MAQTMSDD194 pKa = 3.73GEE196 pKa = 4.75SYY198 pKa = 9.99WEE200 pKa = 4.14DD201 pKa = 2.92KK202 pKa = 10.56RR203 pKa = 11.84YY204 pKa = 8.87EE205 pKa = 4.22TMGPVQGDD213 pKa = 3.38NNYY216 pKa = 9.96CDD218 pKa = 3.5QHH220 pKa = 8.16NGDD223 pKa = 3.75VDD225 pKa = 4.33CNKK228 pKa = 10.2HH229 pKa = 5.8LGSDD233 pKa = 4.0GEE235 pKa = 4.36HH236 pKa = 6.2TFRR239 pKa = 11.84VLKK242 pKa = 10.72DD243 pKa = 3.52EE244 pKa = 4.81SNTPVLHH251 pKa = 7.15LGTCKK256 pKa = 10.1TPVYY260 pKa = 10.46GFGYY264 pKa = 9.64EE265 pKa = 3.97DD266 pKa = 3.75VIVFSEE272 pKa = 4.18IEE274 pKa = 3.72KK275 pKa = 9.22DD276 pKa = 3.55TRR278 pKa = 11.84LSCPNCHH285 pKa = 6.2TSCDD289 pKa = 4.02SEE291 pKa = 4.81SMSIVVPEE299 pKa = 3.89ATKK302 pKa = 10.93KK303 pKa = 10.18NVRR306 pKa = 11.84ICGRR310 pKa = 11.84QNCVLIQSSDD320 pKa = 3.62KK321 pKa = 10.5IIKK324 pKa = 7.65VQRR327 pKa = 11.84DD328 pKa = 3.73FHH330 pKa = 7.66QKK332 pKa = 8.21ITDD335 pKa = 3.27EE336 pKa = 4.05RR337 pKa = 11.84VIFEE341 pKa = 4.2VSDD344 pKa = 3.67DD345 pKa = 3.88EE346 pKa = 4.44KK347 pKa = 10.39TYY349 pKa = 10.8SYY351 pKa = 11.32SLTQSCPVVEE361 pKa = 4.0ICEE364 pKa = 5.25AIDD367 pKa = 3.35CHH369 pKa = 7.66YY370 pKa = 11.49CMIRR374 pKa = 11.84LYY376 pKa = 11.14NPTCYY381 pKa = 10.08EE382 pKa = 3.61WFHH385 pKa = 6.17WVMVAGILYY394 pKa = 8.7TLVIFFGLLLLMLKK408 pKa = 10.15PIIKK412 pKa = 10.05LVWFLICIIYY422 pKa = 9.71KK423 pKa = 10.12IIMKK427 pKa = 10.33LSGIMIGKK435 pKa = 8.62SKK437 pKa = 10.7KK438 pKa = 10.44GFSAVNKK445 pKa = 9.85FADD448 pKa = 4.11EE449 pKa = 4.11EE450 pKa = 4.97SSMEE454 pKa = 3.87NLIYY458 pKa = 10.64KK459 pKa = 7.18PTSIVKK465 pKa = 9.89KK466 pKa = 8.87NKK468 pKa = 9.02PMKK471 pKa = 10.16PMIFIILILGLIVTICEE488 pKa = 4.25SKK490 pKa = 10.39LCSNVVIDD498 pKa = 4.93TIPSQVCTKK507 pKa = 10.44IGDD510 pKa = 3.58QYY512 pKa = 10.06RR513 pKa = 11.84CKK515 pKa = 9.91IGSVVEE521 pKa = 4.18VPMISYY527 pKa = 9.96DD528 pKa = 4.19QISCLHH534 pKa = 6.73FISPKK539 pKa = 10.29GNTIGEE545 pKa = 4.17LQITPMDD552 pKa = 4.49LGFKK556 pKa = 9.86CLKK559 pKa = 9.41EE560 pKa = 3.87NLYY563 pKa = 8.59FTRR566 pKa = 11.84DD567 pKa = 3.35VKK569 pKa = 10.79FHH571 pKa = 6.33VDD573 pKa = 2.92HH574 pKa = 7.54MIRR577 pKa = 11.84CPHH580 pKa = 6.8AGEE583 pKa = 5.96CSDD586 pKa = 4.5DD587 pKa = 3.19WCKK590 pKa = 9.69TVVRR594 pKa = 11.84EE595 pKa = 4.24TKK597 pKa = 10.53VEE599 pKa = 3.98GLSEE603 pKa = 4.24LKK605 pKa = 10.81GMYY608 pKa = 9.66DD609 pKa = 3.28QSCKK613 pKa = 10.54LGSACAGAGCFFCTNSCHH631 pKa = 4.7TTRR634 pKa = 11.84YY635 pKa = 9.77RR636 pKa = 11.84PVPVSGSVYY645 pKa = 10.34EE646 pKa = 4.85IFYY649 pKa = 9.59CPKK652 pKa = 9.01WEE654 pKa = 4.15PSGTFHH660 pKa = 7.59FEE662 pKa = 3.91WKK664 pKa = 10.34GSASTVSTTLNLIHH678 pKa = 6.54GQTVTISADD687 pKa = 2.87IRR689 pKa = 11.84ATLQMTVQAKK699 pKa = 10.52LPVLGKK705 pKa = 10.8KK706 pKa = 10.46FMQKK710 pKa = 10.38DD711 pKa = 3.34QLLAVIDD718 pKa = 4.37GAAKK722 pKa = 9.73NQPQVGVIGQIQCPTVKK739 pKa = 10.44DD740 pKa = 4.19AIEE743 pKa = 4.19MSDD746 pKa = 3.72NCLMAPNSCQCNPTGDD762 pKa = 4.25TDD764 pKa = 4.06SCDD767 pKa = 3.57CSEE770 pKa = 4.3VSLASGFQSEE780 pKa = 4.62NQLPLHH786 pKa = 6.67IGGDD790 pKa = 4.11YY791 pKa = 9.34LTQARR796 pKa = 11.84DD797 pKa = 3.45IPVLKK802 pKa = 10.67ASAYY806 pKa = 8.61GSALIRR812 pKa = 11.84VHH814 pKa = 6.33ATGTLISDD822 pKa = 3.7YY823 pKa = 11.38AEE825 pKa = 4.12EE826 pKa = 4.46EE827 pKa = 4.11EE828 pKa = 5.27CKK830 pKa = 9.65ITMGKK835 pKa = 10.37LNGCHH840 pKa = 6.09SCITGANLTYY850 pKa = 10.08TCNTAKK856 pKa = 10.09PASVVMKK863 pKa = 10.77CGGDD867 pKa = 3.32LNLILDD873 pKa = 4.93CDD875 pKa = 4.09SQDD878 pKa = 3.22KK879 pKa = 10.5QRR881 pKa = 11.84VARR884 pKa = 11.84VHH886 pKa = 6.22IPSPIIDD893 pKa = 3.78VACQTTCSKK902 pKa = 10.92EE903 pKa = 3.9KK904 pKa = 10.73LRR906 pKa = 11.84LTGVLKK912 pKa = 10.85SVGLSAVNNGSHH924 pKa = 6.15IQLIPSEE931 pKa = 4.4SMTDD935 pKa = 3.08LGSWLKK941 pKa = 10.91SIWAYY946 pKa = 9.6FSWWYY951 pKa = 8.72TFVVALIILMIACLLLYY968 pKa = 10.17GFHH971 pKa = 6.87KK972 pKa = 10.64FKK974 pKa = 11.03VAFIEE979 pKa = 4.15KK980 pKa = 9.61QKK982 pKa = 11.16VYY984 pKa = 11.12
MM1 pKa = 7.68ASVKK5 pKa = 10.42LLFVIYY11 pKa = 9.81CISSLMINEE20 pKa = 3.94MRR22 pKa = 11.84TVEE25 pKa = 5.16LPEE28 pKa = 3.97FWHH31 pKa = 6.3MSRR34 pKa = 11.84EE35 pKa = 3.94KK36 pKa = 10.35MNRR39 pKa = 11.84KK40 pKa = 9.26PIDD43 pKa = 3.64PNVKK47 pKa = 10.01VKK49 pKa = 10.79GGNCSNGVPANATIQDD65 pKa = 3.61EE66 pKa = 5.17GITLNKK72 pKa = 9.82CSKK75 pKa = 10.75QMTAVVRR82 pKa = 11.84YY83 pKa = 6.61TVEE86 pKa = 4.23GVDD89 pKa = 3.36EE90 pKa = 4.41TFGCVVNYY98 pKa = 10.61DD99 pKa = 3.88CGLNLHH105 pKa = 6.63NGDD108 pKa = 4.82GYY110 pKa = 10.23CTLKK114 pKa = 11.15NEE116 pKa = 4.62DD117 pKa = 4.34CFHH120 pKa = 6.88EE121 pKa = 4.81PGCIVPKK128 pKa = 10.7DD129 pKa = 4.06RR130 pKa = 11.84ILPMGNCNEE139 pKa = 4.34THH141 pKa = 5.39WTQNRR146 pKa = 11.84PEE148 pKa = 4.06FCSVGVHH155 pKa = 6.38KK156 pKa = 10.72AGCEE160 pKa = 3.62EE161 pKa = 3.97TGRR164 pKa = 11.84VVRR167 pKa = 11.84LPWVVDD173 pKa = 3.72RR174 pKa = 11.84SGKK177 pKa = 9.74KK178 pKa = 9.96RR179 pKa = 11.84LINKK183 pKa = 9.23LNIEE187 pKa = 4.39MAQTMSDD194 pKa = 3.73GEE196 pKa = 4.75SYY198 pKa = 9.99WEE200 pKa = 4.14DD201 pKa = 2.92KK202 pKa = 10.56RR203 pKa = 11.84YY204 pKa = 8.87EE205 pKa = 4.22TMGPVQGDD213 pKa = 3.38NNYY216 pKa = 9.96CDD218 pKa = 3.5QHH220 pKa = 8.16NGDD223 pKa = 3.75VDD225 pKa = 4.33CNKK228 pKa = 10.2HH229 pKa = 5.8LGSDD233 pKa = 4.0GEE235 pKa = 4.36HH236 pKa = 6.2TFRR239 pKa = 11.84VLKK242 pKa = 10.72DD243 pKa = 3.52EE244 pKa = 4.81SNTPVLHH251 pKa = 7.15LGTCKK256 pKa = 10.1TPVYY260 pKa = 10.46GFGYY264 pKa = 9.64EE265 pKa = 3.97DD266 pKa = 3.75VIVFSEE272 pKa = 4.18IEE274 pKa = 3.72KK275 pKa = 9.22DD276 pKa = 3.55TRR278 pKa = 11.84LSCPNCHH285 pKa = 6.2TSCDD289 pKa = 4.02SEE291 pKa = 4.81SMSIVVPEE299 pKa = 3.89ATKK302 pKa = 10.93KK303 pKa = 10.18NVRR306 pKa = 11.84ICGRR310 pKa = 11.84QNCVLIQSSDD320 pKa = 3.62KK321 pKa = 10.5IIKK324 pKa = 7.65VQRR327 pKa = 11.84DD328 pKa = 3.73FHH330 pKa = 7.66QKK332 pKa = 8.21ITDD335 pKa = 3.27EE336 pKa = 4.05RR337 pKa = 11.84VIFEE341 pKa = 4.2VSDD344 pKa = 3.67DD345 pKa = 3.88EE346 pKa = 4.44KK347 pKa = 10.39TYY349 pKa = 10.8SYY351 pKa = 11.32SLTQSCPVVEE361 pKa = 4.0ICEE364 pKa = 5.25AIDD367 pKa = 3.35CHH369 pKa = 7.66YY370 pKa = 11.49CMIRR374 pKa = 11.84LYY376 pKa = 11.14NPTCYY381 pKa = 10.08EE382 pKa = 3.61WFHH385 pKa = 6.17WVMVAGILYY394 pKa = 8.7TLVIFFGLLLLMLKK408 pKa = 10.15PIIKK412 pKa = 10.05LVWFLICIIYY422 pKa = 9.71KK423 pKa = 10.12IIMKK427 pKa = 10.33LSGIMIGKK435 pKa = 8.62SKK437 pKa = 10.7KK438 pKa = 10.44GFSAVNKK445 pKa = 9.85FADD448 pKa = 4.11EE449 pKa = 4.11EE450 pKa = 4.97SSMEE454 pKa = 3.87NLIYY458 pKa = 10.64KK459 pKa = 7.18PTSIVKK465 pKa = 9.89KK466 pKa = 8.87NKK468 pKa = 9.02PMKK471 pKa = 10.16PMIFIILILGLIVTICEE488 pKa = 4.25SKK490 pKa = 10.39LCSNVVIDD498 pKa = 4.93TIPSQVCTKK507 pKa = 10.44IGDD510 pKa = 3.58QYY512 pKa = 10.06RR513 pKa = 11.84CKK515 pKa = 9.91IGSVVEE521 pKa = 4.18VPMISYY527 pKa = 9.96DD528 pKa = 4.19QISCLHH534 pKa = 6.73FISPKK539 pKa = 10.29GNTIGEE545 pKa = 4.17LQITPMDD552 pKa = 4.49LGFKK556 pKa = 9.86CLKK559 pKa = 9.41EE560 pKa = 3.87NLYY563 pKa = 8.59FTRR566 pKa = 11.84DD567 pKa = 3.35VKK569 pKa = 10.79FHH571 pKa = 6.33VDD573 pKa = 2.92HH574 pKa = 7.54MIRR577 pKa = 11.84CPHH580 pKa = 6.8AGEE583 pKa = 5.96CSDD586 pKa = 4.5DD587 pKa = 3.19WCKK590 pKa = 9.69TVVRR594 pKa = 11.84EE595 pKa = 4.24TKK597 pKa = 10.53VEE599 pKa = 3.98GLSEE603 pKa = 4.24LKK605 pKa = 10.81GMYY608 pKa = 9.66DD609 pKa = 3.28QSCKK613 pKa = 10.54LGSACAGAGCFFCTNSCHH631 pKa = 4.7TTRR634 pKa = 11.84YY635 pKa = 9.77RR636 pKa = 11.84PVPVSGSVYY645 pKa = 10.34EE646 pKa = 4.85IFYY649 pKa = 9.59CPKK652 pKa = 9.01WEE654 pKa = 4.15PSGTFHH660 pKa = 7.59FEE662 pKa = 3.91WKK664 pKa = 10.34GSASTVSTTLNLIHH678 pKa = 6.54GQTVTISADD687 pKa = 2.87IRR689 pKa = 11.84ATLQMTVQAKK699 pKa = 10.52LPVLGKK705 pKa = 10.8KK706 pKa = 10.46FMQKK710 pKa = 10.38DD711 pKa = 3.34QLLAVIDD718 pKa = 4.37GAAKK722 pKa = 9.73NQPQVGVIGQIQCPTVKK739 pKa = 10.44DD740 pKa = 4.19AIEE743 pKa = 4.19MSDD746 pKa = 3.72NCLMAPNSCQCNPTGDD762 pKa = 4.25TDD764 pKa = 4.06SCDD767 pKa = 3.57CSEE770 pKa = 4.3VSLASGFQSEE780 pKa = 4.62NQLPLHH786 pKa = 6.67IGGDD790 pKa = 4.11YY791 pKa = 9.34LTQARR796 pKa = 11.84DD797 pKa = 3.45IPVLKK802 pKa = 10.67ASAYY806 pKa = 8.61GSALIRR812 pKa = 11.84VHH814 pKa = 6.33ATGTLISDD822 pKa = 3.7YY823 pKa = 11.38AEE825 pKa = 4.12EE826 pKa = 4.46EE827 pKa = 4.11EE828 pKa = 5.27CKK830 pKa = 9.65ITMGKK835 pKa = 10.37LNGCHH840 pKa = 6.09SCITGANLTYY850 pKa = 10.08TCNTAKK856 pKa = 10.09PASVVMKK863 pKa = 10.77CGGDD867 pKa = 3.32LNLILDD873 pKa = 4.93CDD875 pKa = 4.09SQDD878 pKa = 3.22KK879 pKa = 10.5QRR881 pKa = 11.84VARR884 pKa = 11.84VHH886 pKa = 6.22IPSPIIDD893 pKa = 3.78VACQTTCSKK902 pKa = 10.92EE903 pKa = 3.9KK904 pKa = 10.73LRR906 pKa = 11.84LTGVLKK912 pKa = 10.85SVGLSAVNNGSHH924 pKa = 6.15IQLIPSEE931 pKa = 4.4SMTDD935 pKa = 3.08LGSWLKK941 pKa = 10.91SIWAYY946 pKa = 9.6FSWWYY951 pKa = 8.72TFVVALIILMIACLLLYY968 pKa = 10.17GFHH971 pKa = 6.87KK972 pKa = 10.64FKK974 pKa = 11.03VAFIEE979 pKa = 4.15KK980 pKa = 9.61QKK982 pKa = 11.16VYY984 pKa = 11.12
Molecular weight: 109.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5VGS1|W5VGS1_9VIRU Replicase OS=Cumuto virus OX=1457166 PE=4 SV=1
MM1 pKa = 7.49SIVPTVDD8 pKa = 3.32DD9 pKa = 3.53QLVAEE14 pKa = 5.58GIISDD19 pKa = 4.91LADD22 pKa = 3.86DD23 pKa = 4.99VINDD27 pKa = 3.21STLAVKK33 pKa = 10.29SINLAYY39 pKa = 10.45QGFDD43 pKa = 3.15PVYY46 pKa = 10.3LMQVLAYY53 pKa = 9.45RR54 pKa = 11.84ARR56 pKa = 11.84DD57 pKa = 3.62AKK59 pKa = 10.58ISAVNHH65 pKa = 5.85KK66 pKa = 11.01SNLRR70 pKa = 11.84TLALIGTMRR79 pKa = 11.84GSKK82 pKa = 9.82IEE84 pKa = 4.33TISGRR89 pKa = 11.84SGQEE93 pKa = 3.48LKK95 pKa = 11.12DD96 pKa = 3.81FLVKK100 pKa = 10.06MVRR103 pKa = 11.84DD104 pKa = 3.85YY105 pKa = 11.2KK106 pKa = 10.84LKK108 pKa = 10.5SGRR111 pKa = 11.84PTSNQDD117 pKa = 2.68ATLLRR122 pKa = 11.84IAAIYY127 pKa = 8.29AAPLAIAIKK136 pKa = 10.09NNAVSITTTIAPNNVAPGYY155 pKa = 9.47PRR157 pKa = 11.84FMCLSTFGALIPSAEE172 pKa = 4.15SMRR175 pKa = 11.84EE176 pKa = 3.93GEE178 pKa = 4.08QALIAGAFAEE188 pKa = 4.41HH189 pKa = 6.1QALFDD194 pKa = 3.67RR195 pKa = 11.84VINSRR200 pKa = 11.84TKK202 pKa = 9.72NYY204 pKa = 10.27SPKK207 pKa = 10.69DD208 pKa = 3.54VIKK211 pKa = 10.83NYY213 pKa = 10.07IQIQQLSALYY223 pKa = 10.66SEE225 pKa = 4.91GDD227 pKa = 3.56RR228 pKa = 11.84IKK230 pKa = 10.14TLLALGLIEE239 pKa = 4.32MMGNQMYY246 pKa = 10.07QVTALYY252 pKa = 10.39RR253 pKa = 11.84SALNAAKK260 pKa = 10.14QKK262 pKa = 9.74WDD264 pKa = 3.25ARR266 pKa = 11.84VV267 pKa = 2.9
MM1 pKa = 7.49SIVPTVDD8 pKa = 3.32DD9 pKa = 3.53QLVAEE14 pKa = 5.58GIISDD19 pKa = 4.91LADD22 pKa = 3.86DD23 pKa = 4.99VINDD27 pKa = 3.21STLAVKK33 pKa = 10.29SINLAYY39 pKa = 10.45QGFDD43 pKa = 3.15PVYY46 pKa = 10.3LMQVLAYY53 pKa = 9.45RR54 pKa = 11.84ARR56 pKa = 11.84DD57 pKa = 3.62AKK59 pKa = 10.58ISAVNHH65 pKa = 5.85KK66 pKa = 11.01SNLRR70 pKa = 11.84TLALIGTMRR79 pKa = 11.84GSKK82 pKa = 9.82IEE84 pKa = 4.33TISGRR89 pKa = 11.84SGQEE93 pKa = 3.48LKK95 pKa = 11.12DD96 pKa = 3.81FLVKK100 pKa = 10.06MVRR103 pKa = 11.84DD104 pKa = 3.85YY105 pKa = 11.2KK106 pKa = 10.84LKK108 pKa = 10.5SGRR111 pKa = 11.84PTSNQDD117 pKa = 2.68ATLLRR122 pKa = 11.84IAAIYY127 pKa = 8.29AAPLAIAIKK136 pKa = 10.09NNAVSITTTIAPNNVAPGYY155 pKa = 9.47PRR157 pKa = 11.84FMCLSTFGALIPSAEE172 pKa = 4.15SMRR175 pKa = 11.84EE176 pKa = 3.93GEE178 pKa = 4.08QALIAGAFAEE188 pKa = 4.41HH189 pKa = 6.1QALFDD194 pKa = 3.67RR195 pKa = 11.84VINSRR200 pKa = 11.84TKK202 pKa = 9.72NYY204 pKa = 10.27SPKK207 pKa = 10.69DD208 pKa = 3.54VIKK211 pKa = 10.83NYY213 pKa = 10.07IQIQQLSALYY223 pKa = 10.66SEE225 pKa = 4.91GDD227 pKa = 3.56RR228 pKa = 11.84IKK230 pKa = 10.14TLLALGLIEE239 pKa = 4.32MMGNQMYY246 pKa = 10.07QVTALYY252 pKa = 10.39RR253 pKa = 11.84SALNAAKK260 pKa = 10.14QKK262 pKa = 9.74WDD264 pKa = 3.25ARR266 pKa = 11.84VV267 pKa = 2.9
Molecular weight: 29.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3322 |
267 |
2071 |
1107.3 |
126.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.877 ± 1.932 | 2.348 ± 1.511 |
5.81 ± 0.268 | 6.893 ± 1.277 |
3.793 ± 0.504 | 5.027 ± 0.819 |
1.836 ± 0.422 | 7.164 ± 0.519 |
7.556 ± 0.602 | 9.482 ± 0.864 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.402 ± 0.122 | 4.846 ± 0.185 |
3.402 ± 0.315 | 3.642 ± 0.316 |
4.967 ± 0.891 | 8.158 ± 0.477 |
5.418 ± 0.317 | 6.412 ± 0.754 |
1.595 ± 0.353 | 3.371 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
