
Sendai virus (strain Z) (SeV) (Sendai virus (strain HVJ))
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Respirovirus; Murine respirovirus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
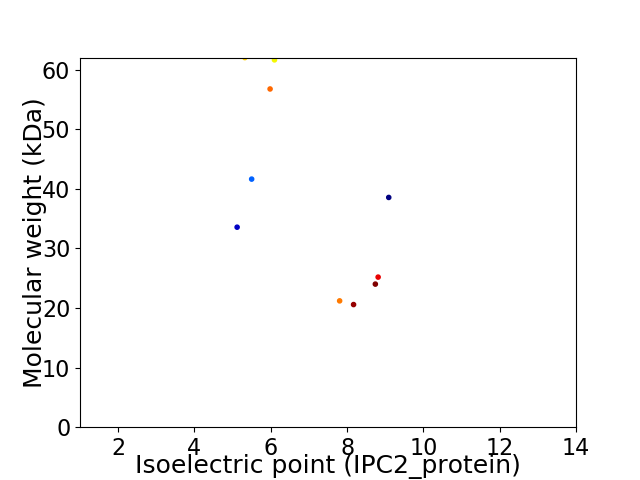
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P04862-2|C-2_SENDZ Isoform of P04862 Isoform C protein of C' protein OS=Sendai virus (strain Z) OX=11198 GN=P/V/C PE=1 SV=3
MM1 pKa = 7.56DD2 pKa = 3.68QDD4 pKa = 4.41AFILKK9 pKa = 9.71EE10 pKa = 3.88DD11 pKa = 4.02SEE13 pKa = 4.98VEE15 pKa = 3.95RR16 pKa = 11.84EE17 pKa = 4.15APGGRR22 pKa = 11.84EE23 pKa = 3.99SLSDD27 pKa = 3.58VIGFLDD33 pKa = 3.71AVLSSEE39 pKa = 4.09PTDD42 pKa = 3.29IGGDD46 pKa = 3.55RR47 pKa = 11.84SWLHH51 pKa = 5.09NTINTPQGPGSAHH64 pKa = 6.25RR65 pKa = 11.84AKK67 pKa = 10.78SEE69 pKa = 4.15GEE71 pKa = 4.42GEE73 pKa = 4.28VSTPSTQDD81 pKa = 2.88NRR83 pKa = 11.84SGEE86 pKa = 4.16EE87 pKa = 3.75SRR89 pKa = 11.84VSGRR93 pKa = 11.84TSKK96 pKa = 11.09PEE98 pKa = 3.94AEE100 pKa = 3.93AHH102 pKa = 6.43AGNLDD107 pKa = 3.26KK108 pKa = 11.5QNIHH112 pKa = 6.5RR113 pKa = 11.84AFGGRR118 pKa = 11.84TGTNSVSQDD127 pKa = 3.37LGDD130 pKa = 4.65GGDD133 pKa = 3.67SGILEE138 pKa = 4.43NPPNEE143 pKa = 3.91RR144 pKa = 11.84GYY146 pKa = 9.84PRR148 pKa = 11.84SGIEE152 pKa = 3.92DD153 pKa = 3.78EE154 pKa = 4.2NRR156 pKa = 11.84EE157 pKa = 4.11MAAHH161 pKa = 6.69PDD163 pKa = 3.54KK164 pKa = 11.08RR165 pKa = 11.84GEE167 pKa = 4.12DD168 pKa = 3.56QAEE171 pKa = 4.07GLPEE175 pKa = 4.03EE176 pKa = 4.6VRR178 pKa = 11.84GSTSLPDD185 pKa = 3.6EE186 pKa = 4.86GEE188 pKa = 4.37GGASNNGRR196 pKa = 11.84SMEE199 pKa = 4.61PGSSHH204 pKa = 5.66SARR207 pKa = 11.84VTGVLVIPSPEE218 pKa = 3.91LEE220 pKa = 4.07EE221 pKa = 4.42AVLRR225 pKa = 11.84RR226 pKa = 11.84NKK228 pKa = 9.86RR229 pKa = 11.84RR230 pKa = 11.84PTNSGSKK237 pKa = 9.48PLTPATVPGTRR248 pKa = 11.84SPPLNRR254 pKa = 11.84YY255 pKa = 8.99NSTGSPPGKK264 pKa = 9.58PPSTQDD270 pKa = 2.94EE271 pKa = 5.06HH272 pKa = 8.35INSGDD277 pKa = 3.51TPAVRR282 pKa = 11.84VKK284 pKa = 10.65DD285 pKa = 4.02RR286 pKa = 11.84KK287 pKa = 9.93PPIGTRR293 pKa = 11.84SVSDD297 pKa = 3.76CPANGRR303 pKa = 11.84SIHH306 pKa = 6.83PGLEE310 pKa = 3.79TDD312 pKa = 3.74STKK315 pKa = 10.79KK316 pKa = 10.53GAA318 pKa = 3.75
MM1 pKa = 7.56DD2 pKa = 3.68QDD4 pKa = 4.41AFILKK9 pKa = 9.71EE10 pKa = 3.88DD11 pKa = 4.02SEE13 pKa = 4.98VEE15 pKa = 3.95RR16 pKa = 11.84EE17 pKa = 4.15APGGRR22 pKa = 11.84EE23 pKa = 3.99SLSDD27 pKa = 3.58VIGFLDD33 pKa = 3.71AVLSSEE39 pKa = 4.09PTDD42 pKa = 3.29IGGDD46 pKa = 3.55RR47 pKa = 11.84SWLHH51 pKa = 5.09NTINTPQGPGSAHH64 pKa = 6.25RR65 pKa = 11.84AKK67 pKa = 10.78SEE69 pKa = 4.15GEE71 pKa = 4.42GEE73 pKa = 4.28VSTPSTQDD81 pKa = 2.88NRR83 pKa = 11.84SGEE86 pKa = 4.16EE87 pKa = 3.75SRR89 pKa = 11.84VSGRR93 pKa = 11.84TSKK96 pKa = 11.09PEE98 pKa = 3.94AEE100 pKa = 3.93AHH102 pKa = 6.43AGNLDD107 pKa = 3.26KK108 pKa = 11.5QNIHH112 pKa = 6.5RR113 pKa = 11.84AFGGRR118 pKa = 11.84TGTNSVSQDD127 pKa = 3.37LGDD130 pKa = 4.65GGDD133 pKa = 3.67SGILEE138 pKa = 4.43NPPNEE143 pKa = 3.91RR144 pKa = 11.84GYY146 pKa = 9.84PRR148 pKa = 11.84SGIEE152 pKa = 3.92DD153 pKa = 3.78EE154 pKa = 4.2NRR156 pKa = 11.84EE157 pKa = 4.11MAAHH161 pKa = 6.69PDD163 pKa = 3.54KK164 pKa = 11.08RR165 pKa = 11.84GEE167 pKa = 4.12DD168 pKa = 3.56QAEE171 pKa = 4.07GLPEE175 pKa = 4.03EE176 pKa = 4.6VRR178 pKa = 11.84GSTSLPDD185 pKa = 3.6EE186 pKa = 4.86GEE188 pKa = 4.37GGASNNGRR196 pKa = 11.84SMEE199 pKa = 4.61PGSSHH204 pKa = 5.66SARR207 pKa = 11.84VTGVLVIPSPEE218 pKa = 3.91LEE220 pKa = 4.07EE221 pKa = 4.42AVLRR225 pKa = 11.84RR226 pKa = 11.84NKK228 pKa = 9.86RR229 pKa = 11.84RR230 pKa = 11.84PTNSGSKK237 pKa = 9.48PLTPATVPGTRR248 pKa = 11.84SPPLNRR254 pKa = 11.84YY255 pKa = 8.99NSTGSPPGKK264 pKa = 9.58PPSTQDD270 pKa = 2.94EE271 pKa = 5.06HH272 pKa = 8.35INSGDD277 pKa = 3.51TPAVRR282 pKa = 11.84VKK284 pKa = 10.65DD285 pKa = 4.02RR286 pKa = 11.84KK287 pKa = 9.93PPIGTRR293 pKa = 11.84SVSDD297 pKa = 3.76CPANGRR303 pKa = 11.84SIHH306 pKa = 6.83PGLEE310 pKa = 3.79TDD312 pKa = 3.74STKK315 pKa = 10.79KK316 pKa = 10.53GAA318 pKa = 3.75
Molecular weight: 33.57 kDa
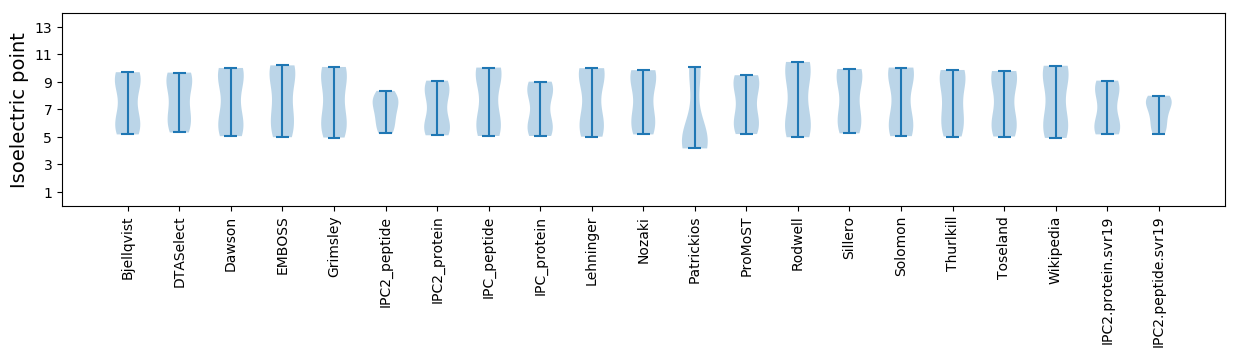
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P69282|V_SENDZ Protein V OS=Sendai virus (strain Z) OX=11198 GN=P/V/C PE=1 SV=1
MM1 pKa = 7.6ADD3 pKa = 3.36IYY5 pKa = 10.85RR6 pKa = 11.84FPKK9 pKa = 10.59FSYY12 pKa = 9.74EE13 pKa = 4.12DD14 pKa = 3.44NGTVEE19 pKa = 4.59PLPLRR24 pKa = 11.84TGPDD28 pKa = 3.01KK29 pKa = 10.99KK30 pKa = 10.56AIPHH34 pKa = 5.1IRR36 pKa = 11.84IVKK39 pKa = 10.01VGDD42 pKa = 3.63PPKK45 pKa = 10.69HH46 pKa = 4.84GVRR49 pKa = 11.84YY50 pKa = 10.03LDD52 pKa = 3.96LLLLGFFEE60 pKa = 4.98TPKK63 pKa = 9.37QTTNLGSVSDD73 pKa = 3.87LTEE76 pKa = 3.94PTSYY80 pKa = 10.56SICGSGSLPIGVAKK94 pKa = 10.76YY95 pKa = 10.52YY96 pKa = 9.98GTDD99 pKa = 3.29QEE101 pKa = 4.62LLKK104 pKa = 11.01ACTDD108 pKa = 3.21LRR110 pKa = 11.84ITVRR114 pKa = 11.84RR115 pKa = 11.84TVRR118 pKa = 11.84AGEE121 pKa = 4.17MIVYY125 pKa = 8.62MVDD128 pKa = 3.46SIGAPLLPWSGRR140 pKa = 11.84LRR142 pKa = 11.84QGMIFNANKK151 pKa = 9.77VALAPQCLPVDD162 pKa = 3.24KK163 pKa = 10.48DD164 pKa = 2.85IRR166 pKa = 11.84LRR168 pKa = 11.84VVFVNGTSLGAITIAKK184 pKa = 9.25IPKK187 pKa = 8.6TLADD191 pKa = 3.79LALPNSISVNLLVTLKK207 pKa = 10.0TGISTEE213 pKa = 4.15QKK215 pKa = 10.47GVLPVLDD222 pKa = 4.08DD223 pKa = 3.73QGEE226 pKa = 4.28KK227 pKa = 10.49KK228 pKa = 10.88LNFMVHH234 pKa = 6.52LGLIRR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.27VGKK244 pKa = 9.46IYY246 pKa = 10.67SVEE249 pKa = 4.02YY250 pKa = 9.84CKK252 pKa = 11.04SKK254 pKa = 10.18IEE256 pKa = 3.93RR257 pKa = 11.84MRR259 pKa = 11.84LIFSLGLIGGISFHH273 pKa = 5.57VQVNGTLSKK282 pKa = 10.14TFMSQLAWKK291 pKa = 10.01RR292 pKa = 11.84AVCFPLMDD300 pKa = 4.24VNPHH304 pKa = 4.81MNMVIWAASVEE315 pKa = 4.3ITGVDD320 pKa = 3.49AVFQPAIPRR329 pKa = 11.84DD330 pKa = 3.38FRR332 pKa = 11.84YY333 pKa = 10.38YY334 pKa = 10.21PNVVAKK340 pKa = 10.68NIGRR344 pKa = 11.84IRR346 pKa = 11.84KK347 pKa = 8.53LL348 pKa = 3.16
MM1 pKa = 7.6ADD3 pKa = 3.36IYY5 pKa = 10.85RR6 pKa = 11.84FPKK9 pKa = 10.59FSYY12 pKa = 9.74EE13 pKa = 4.12DD14 pKa = 3.44NGTVEE19 pKa = 4.59PLPLRR24 pKa = 11.84TGPDD28 pKa = 3.01KK29 pKa = 10.99KK30 pKa = 10.56AIPHH34 pKa = 5.1IRR36 pKa = 11.84IVKK39 pKa = 10.01VGDD42 pKa = 3.63PPKK45 pKa = 10.69HH46 pKa = 4.84GVRR49 pKa = 11.84YY50 pKa = 10.03LDD52 pKa = 3.96LLLLGFFEE60 pKa = 4.98TPKK63 pKa = 9.37QTTNLGSVSDD73 pKa = 3.87LTEE76 pKa = 3.94PTSYY80 pKa = 10.56SICGSGSLPIGVAKK94 pKa = 10.76YY95 pKa = 10.52YY96 pKa = 9.98GTDD99 pKa = 3.29QEE101 pKa = 4.62LLKK104 pKa = 11.01ACTDD108 pKa = 3.21LRR110 pKa = 11.84ITVRR114 pKa = 11.84RR115 pKa = 11.84TVRR118 pKa = 11.84AGEE121 pKa = 4.17MIVYY125 pKa = 8.62MVDD128 pKa = 3.46SIGAPLLPWSGRR140 pKa = 11.84LRR142 pKa = 11.84QGMIFNANKK151 pKa = 9.77VALAPQCLPVDD162 pKa = 3.24KK163 pKa = 10.48DD164 pKa = 2.85IRR166 pKa = 11.84LRR168 pKa = 11.84VVFVNGTSLGAITIAKK184 pKa = 9.25IPKK187 pKa = 8.6TLADD191 pKa = 3.79LALPNSISVNLLVTLKK207 pKa = 10.0TGISTEE213 pKa = 4.15QKK215 pKa = 10.47GVLPVLDD222 pKa = 4.08DD223 pKa = 3.73QGEE226 pKa = 4.28KK227 pKa = 10.49KK228 pKa = 10.88LNFMVHH234 pKa = 6.52LGLIRR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.27VGKK244 pKa = 9.46IYY246 pKa = 10.67SVEE249 pKa = 4.02YY250 pKa = 9.84CKK252 pKa = 11.04SKK254 pKa = 10.18IEE256 pKa = 3.93RR257 pKa = 11.84MRR259 pKa = 11.84LIFSLGLIGGISFHH273 pKa = 5.57VQVNGTLSKK282 pKa = 10.14TFMSQLAWKK291 pKa = 10.01RR292 pKa = 11.84AVCFPLMDD300 pKa = 4.24VNPHH304 pKa = 4.81MNMVIWAASVEE315 pKa = 4.3ITGVDD320 pKa = 3.49AVFQPAIPRR329 pKa = 11.84DD330 pKa = 3.38FRR332 pKa = 11.84YY333 pKa = 10.38YY334 pKa = 10.21PNVVAKK340 pKa = 10.68NIGRR344 pKa = 11.84IRR346 pKa = 11.84KK347 pKa = 8.53LL348 pKa = 3.16
Molecular weight: 38.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3475 |
175 |
568 |
347.5 |
38.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.216 ± 0.587 | 1.065 ± 0.286 |
5.295 ± 0.412 | 8.144 ± 1.137 |
1.957 ± 0.407 | 7.453 ± 0.844 |
1.871 ± 0.161 | 6.129 ± 0.866 |
5.928 ± 0.618 | 8.863 ± 0.88 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.561 ± 0.403 | 3.54 ± 0.482 |
5.468 ± 0.802 | 3.914 ± 0.553 |
7.367 ± 0.511 | 8.489 ± 0.923 |
6.647 ± 0.269 | 6.187 ± 0.631 |
0.806 ± 0.181 | 2.101 ± 0.314 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
