
HCBI9.212 virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykrogvirus; Gemykrogvirus bovas1; Bovine associated gemykrogvirus 1
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
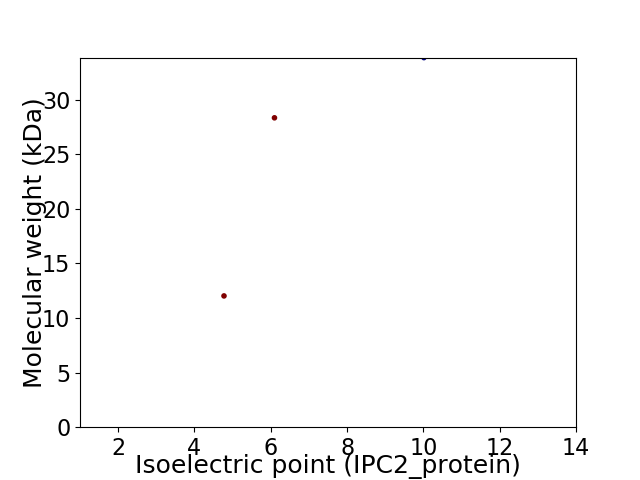
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A077XNA4|A0A077XNA4_9VIRU Replication associated protein OS=HCBI9.212 virus OX=1516080 GN=rep PE=4 SV=1
MM1 pKa = 8.18DD2 pKa = 5.2EE3 pKa = 5.56FSDD6 pKa = 3.8DD7 pKa = 3.12CQYY10 pKa = 12.03AVFDD14 pKa = 5.26DD15 pKa = 3.78IQGGFEE21 pKa = 4.13YY22 pKa = 10.4FHH24 pKa = 7.24SYY26 pKa = 10.2KK27 pKa = 10.66GWLGAQKK34 pKa = 10.48QFVITDD40 pKa = 3.92KK41 pKa = 10.7YY42 pKa = 9.41RR43 pKa = 11.84KK44 pKa = 9.57KK45 pKa = 10.0RR46 pKa = 11.84TIYY49 pKa = 8.85WGKK52 pKa = 9.5PSIMCMNEE60 pKa = 3.78DD61 pKa = 3.13PHH63 pKa = 6.07FAKK66 pKa = 10.77GVDD69 pKa = 3.94YY70 pKa = 10.96DD71 pKa = 3.44WLMNNCHH78 pKa = 7.1IINVVDD84 pKa = 4.89PICIVSNPPSSNEE97 pKa = 4.04SQSEE101 pKa = 4.24QII103 pKa = 4.15
MM1 pKa = 8.18DD2 pKa = 5.2EE3 pKa = 5.56FSDD6 pKa = 3.8DD7 pKa = 3.12CQYY10 pKa = 12.03AVFDD14 pKa = 5.26DD15 pKa = 3.78IQGGFEE21 pKa = 4.13YY22 pKa = 10.4FHH24 pKa = 7.24SYY26 pKa = 10.2KK27 pKa = 10.66GWLGAQKK34 pKa = 10.48QFVITDD40 pKa = 3.92KK41 pKa = 10.7YY42 pKa = 9.41RR43 pKa = 11.84KK44 pKa = 9.57KK45 pKa = 10.0RR46 pKa = 11.84TIYY49 pKa = 8.85WGKK52 pKa = 9.5PSIMCMNEE60 pKa = 3.78DD61 pKa = 3.13PHH63 pKa = 6.07FAKK66 pKa = 10.77GVDD69 pKa = 3.94YY70 pKa = 10.96DD71 pKa = 3.44WLMNNCHH78 pKa = 7.1IINVVDD84 pKa = 4.89PICIVSNPPSSNEE97 pKa = 4.04SQSEE101 pKa = 4.24QII103 pKa = 4.15
Molecular weight: 12.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077XLW1|A0A077XLW1_9VIRU Replication protein OS=HCBI9.212 virus OX=1516080 GN=Rep PE=3 SV=1
MM1 pKa = 7.83AYY3 pKa = 10.26GFRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FGRR11 pKa = 11.84SRR13 pKa = 11.84RR14 pKa = 11.84SYY16 pKa = 9.2RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84PVRR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 8.9SRR26 pKa = 11.84RR27 pKa = 11.84GRR29 pKa = 11.84SGYY32 pKa = 10.52GRR34 pKa = 11.84GGYY37 pKa = 7.78NRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 9.23RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84SWISPYY53 pKa = 10.49NKK55 pKa = 9.56KK56 pKa = 10.1HH57 pKa = 6.05DD58 pKa = 4.48VIKK61 pKa = 10.53GAHH64 pKa = 6.95DD65 pKa = 4.0SNDD68 pKa = 3.11NSFAPIGAPNTFFGFCPTYY87 pKa = 10.07MPNAVAGDD95 pKa = 3.86TGHH98 pKa = 6.52NPQKK102 pKa = 10.72VRR104 pKa = 11.84TSKK107 pKa = 10.64NVGFSGYY114 pKa = 7.18QEE116 pKa = 3.84RR117 pKa = 11.84VHH119 pKa = 6.45IATAEE124 pKa = 4.03SLIWRR129 pKa = 11.84RR130 pKa = 11.84MVIWSYY136 pKa = 11.7NRR138 pKa = 11.84LEE140 pKa = 4.22NTAGPTKK147 pKa = 10.39TDD149 pKa = 3.57DD150 pKa = 3.17QGTSYY155 pKa = 7.46TTRR158 pKa = 11.84QLTPLQLNEE167 pKa = 3.98GVRR170 pKa = 11.84AFLFRR175 pKa = 11.84GTQGIDD181 pKa = 3.19YY182 pKa = 9.88TEE184 pKa = 4.25NTLHH188 pKa = 6.46EE189 pKa = 4.74APLNSDD195 pKa = 4.15HH196 pKa = 6.53FTVAYY201 pKa = 10.3DD202 pKa = 3.51RR203 pKa = 11.84TRR205 pKa = 11.84VLQPRR210 pKa = 11.84RR211 pKa = 11.84PTGDD215 pKa = 2.99EE216 pKa = 3.82YY217 pKa = 12.03GHH219 pKa = 6.44IFNMKK224 pKa = 8.4FWNPGGRR231 pKa = 11.84IIYY234 pKa = 10.4SDD236 pKa = 5.34DD237 pKa = 3.44EE238 pKa = 6.41DD239 pKa = 4.55GLQKK243 pKa = 11.07GPLVNGWSSLGRR255 pKa = 11.84VSKK258 pKa = 11.21GNMYY262 pKa = 10.44IFDD265 pKa = 4.18VFSTGYY271 pKa = 9.78SLSSNTPSIARR282 pKa = 11.84FAPTGTRR289 pKa = 11.84YY290 pKa = 6.29WTEE293 pKa = 3.8GG294 pKa = 3.14
MM1 pKa = 7.83AYY3 pKa = 10.26GFRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FGRR11 pKa = 11.84SRR13 pKa = 11.84RR14 pKa = 11.84SYY16 pKa = 9.2RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84PVRR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 8.9SRR26 pKa = 11.84RR27 pKa = 11.84GRR29 pKa = 11.84SGYY32 pKa = 10.52GRR34 pKa = 11.84GGYY37 pKa = 7.78NRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 9.23RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84SWISPYY53 pKa = 10.49NKK55 pKa = 9.56KK56 pKa = 10.1HH57 pKa = 6.05DD58 pKa = 4.48VIKK61 pKa = 10.53GAHH64 pKa = 6.95DD65 pKa = 4.0SNDD68 pKa = 3.11NSFAPIGAPNTFFGFCPTYY87 pKa = 10.07MPNAVAGDD95 pKa = 3.86TGHH98 pKa = 6.52NPQKK102 pKa = 10.72VRR104 pKa = 11.84TSKK107 pKa = 10.64NVGFSGYY114 pKa = 7.18QEE116 pKa = 3.84RR117 pKa = 11.84VHH119 pKa = 6.45IATAEE124 pKa = 4.03SLIWRR129 pKa = 11.84RR130 pKa = 11.84MVIWSYY136 pKa = 11.7NRR138 pKa = 11.84LEE140 pKa = 4.22NTAGPTKK147 pKa = 10.39TDD149 pKa = 3.57DD150 pKa = 3.17QGTSYY155 pKa = 7.46TTRR158 pKa = 11.84QLTPLQLNEE167 pKa = 3.98GVRR170 pKa = 11.84AFLFRR175 pKa = 11.84GTQGIDD181 pKa = 3.19YY182 pKa = 9.88TEE184 pKa = 4.25NTLHH188 pKa = 6.46EE189 pKa = 4.74APLNSDD195 pKa = 4.15HH196 pKa = 6.53FTVAYY201 pKa = 10.3DD202 pKa = 3.51RR203 pKa = 11.84TRR205 pKa = 11.84VLQPRR210 pKa = 11.84RR211 pKa = 11.84PTGDD215 pKa = 2.99EE216 pKa = 3.82YY217 pKa = 12.03GHH219 pKa = 6.44IFNMKK224 pKa = 8.4FWNPGGRR231 pKa = 11.84IIYY234 pKa = 10.4SDD236 pKa = 5.34DD237 pKa = 3.44EE238 pKa = 6.41DD239 pKa = 4.55GLQKK243 pKa = 11.07GPLVNGWSSLGRR255 pKa = 11.84VSKK258 pKa = 11.21GNMYY262 pKa = 10.44IFDD265 pKa = 4.18VFSTGYY271 pKa = 9.78SLSSNTPSIARR282 pKa = 11.84FAPTGTRR289 pKa = 11.84YY290 pKa = 6.29WTEE293 pKa = 3.8GG294 pKa = 3.14
Molecular weight: 33.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
641 |
103 |
294 |
213.7 |
24.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.148 ± 0.79 | 1.872 ± 0.94 |
6.24 ± 1.094 | 5.148 ± 1.395 |
4.992 ± 0.28 | 7.956 ± 1.551 |
2.964 ± 0.386 | 6.24 ± 1.307 |
3.9 ± 0.811 | 4.68 ± 0.884 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.872 ± 0.557 | 5.46 ± 0.483 |
5.772 ± 0.405 | 3.744 ± 0.706 |
10.14 ± 2.471 | 6.864 ± 0.768 |
5.148 ± 1.624 | 4.524 ± 0.361 |
1.872 ± 0.363 | 5.46 ± 0.483 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
