
Orungo virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
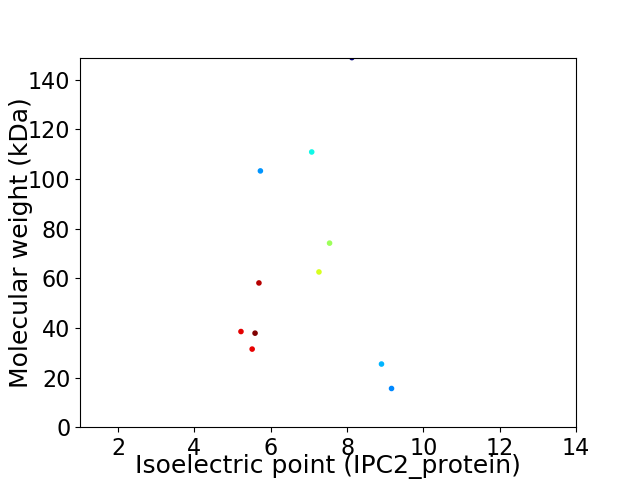
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5QLX5|W5QLX5_9REOV RNA-directed RNA polymerase OS=Orungo virus OX=40058 PE=3 SV=1
MM1 pKa = 6.86AQEE4 pKa = 4.38VKK6 pKa = 10.53RR7 pKa = 11.84KK8 pKa = 6.98FTRR11 pKa = 11.84TVCIYY16 pKa = 10.51DD17 pKa = 3.37PQNRR21 pKa = 11.84TFCGRR26 pKa = 11.84SAAMVCNSYY35 pKa = 8.95YY36 pKa = 8.37TIKK39 pKa = 9.97MGRR42 pKa = 11.84TTQCGVSPVPVPKK55 pKa = 10.32SYY57 pKa = 11.14VLEE60 pKa = 4.73IPDD63 pKa = 3.46VGSYY67 pKa = 10.56RR68 pKa = 11.84ILDD71 pKa = 4.11GQDD74 pKa = 3.49SISVIVTEE82 pKa = 4.53TGVEE86 pKa = 4.05ATSEE90 pKa = 3.87RR91 pKa = 11.84WEE93 pKa = 3.79EE94 pKa = 3.88WKK96 pKa = 10.95FEE98 pKa = 3.93GVNCVPMVVQTEE110 pKa = 4.11IGKK113 pKa = 9.94GLIDD117 pKa = 3.73MEE119 pKa = 4.59IKK121 pKa = 10.48FSKK124 pKa = 10.67GMGLVKK130 pKa = 10.09PYY132 pKa = 10.76VKK134 pKa = 10.76NEE136 pKa = 3.66VSRR139 pKa = 11.84TEE141 pKa = 3.78IPRR144 pKa = 11.84LPGASVIDD152 pKa = 3.87TPIRR156 pKa = 11.84DD157 pKa = 3.6YY158 pKa = 11.24RR159 pKa = 11.84QLLKK163 pKa = 10.63DD164 pKa = 3.3QRR166 pKa = 11.84DD167 pKa = 3.63EE168 pKa = 4.03RR169 pKa = 11.84RR170 pKa = 11.84AALDD174 pKa = 5.02AIACDD179 pKa = 3.32ICPTLSRR186 pKa = 11.84MLGANITDD194 pKa = 4.22GSPQVEE200 pKa = 4.38EE201 pKa = 4.27VATRR205 pKa = 11.84PTQPPAQLKK214 pKa = 10.52LDD216 pKa = 3.86VPNWDD221 pKa = 3.82QPTDD225 pKa = 3.62SDD227 pKa = 5.97DD228 pKa = 3.64EE229 pKa = 4.76DD230 pKa = 4.22EE231 pKa = 4.2EE232 pKa = 4.85SKK234 pKa = 10.97RR235 pKa = 11.84VTTQSNADD243 pKa = 3.54TEE245 pKa = 4.44EE246 pKa = 3.88QVTFEE251 pKa = 5.18DD252 pKa = 5.76CITDD256 pKa = 3.42QYY258 pKa = 11.25FSKK261 pKa = 9.83TGEE264 pKa = 3.94FAKK267 pKa = 10.3KK268 pKa = 9.42YY269 pKa = 9.89PEE271 pKa = 3.95MLAGLSAKK279 pKa = 9.52MPSEE283 pKa = 3.88TGRR286 pKa = 11.84FTKK289 pKa = 10.33ILTTRR294 pKa = 11.84KK295 pKa = 9.36AQWNNVPLFEE305 pKa = 6.23LDD307 pKa = 3.67PNGFTYY313 pKa = 10.48HH314 pKa = 6.68FSKK317 pKa = 10.68LGNSTRR323 pKa = 11.84IFCVQRR329 pKa = 11.84DD330 pKa = 4.06LSYY333 pKa = 10.37MVLPAGQGLKK343 pKa = 10.5
MM1 pKa = 6.86AQEE4 pKa = 4.38VKK6 pKa = 10.53RR7 pKa = 11.84KK8 pKa = 6.98FTRR11 pKa = 11.84TVCIYY16 pKa = 10.51DD17 pKa = 3.37PQNRR21 pKa = 11.84TFCGRR26 pKa = 11.84SAAMVCNSYY35 pKa = 8.95YY36 pKa = 8.37TIKK39 pKa = 9.97MGRR42 pKa = 11.84TTQCGVSPVPVPKK55 pKa = 10.32SYY57 pKa = 11.14VLEE60 pKa = 4.73IPDD63 pKa = 3.46VGSYY67 pKa = 10.56RR68 pKa = 11.84ILDD71 pKa = 4.11GQDD74 pKa = 3.49SISVIVTEE82 pKa = 4.53TGVEE86 pKa = 4.05ATSEE90 pKa = 3.87RR91 pKa = 11.84WEE93 pKa = 3.79EE94 pKa = 3.88WKK96 pKa = 10.95FEE98 pKa = 3.93GVNCVPMVVQTEE110 pKa = 4.11IGKK113 pKa = 9.94GLIDD117 pKa = 3.73MEE119 pKa = 4.59IKK121 pKa = 10.48FSKK124 pKa = 10.67GMGLVKK130 pKa = 10.09PYY132 pKa = 10.76VKK134 pKa = 10.76NEE136 pKa = 3.66VSRR139 pKa = 11.84TEE141 pKa = 3.78IPRR144 pKa = 11.84LPGASVIDD152 pKa = 3.87TPIRR156 pKa = 11.84DD157 pKa = 3.6YY158 pKa = 11.24RR159 pKa = 11.84QLLKK163 pKa = 10.63DD164 pKa = 3.3QRR166 pKa = 11.84DD167 pKa = 3.63EE168 pKa = 4.03RR169 pKa = 11.84RR170 pKa = 11.84AALDD174 pKa = 5.02AIACDD179 pKa = 3.32ICPTLSRR186 pKa = 11.84MLGANITDD194 pKa = 4.22GSPQVEE200 pKa = 4.38EE201 pKa = 4.27VATRR205 pKa = 11.84PTQPPAQLKK214 pKa = 10.52LDD216 pKa = 3.86VPNWDD221 pKa = 3.82QPTDD225 pKa = 3.62SDD227 pKa = 5.97DD228 pKa = 3.64EE229 pKa = 4.76DD230 pKa = 4.22EE231 pKa = 4.2EE232 pKa = 4.85SKK234 pKa = 10.97RR235 pKa = 11.84VTTQSNADD243 pKa = 3.54TEE245 pKa = 4.44EE246 pKa = 3.88QVTFEE251 pKa = 5.18DD252 pKa = 5.76CITDD256 pKa = 3.42QYY258 pKa = 11.25FSKK261 pKa = 9.83TGEE264 pKa = 3.94FAKK267 pKa = 10.3KK268 pKa = 9.42YY269 pKa = 9.89PEE271 pKa = 3.95MLAGLSAKK279 pKa = 9.52MPSEE283 pKa = 3.88TGRR286 pKa = 11.84FTKK289 pKa = 10.33ILTTRR294 pKa = 11.84KK295 pKa = 9.36AQWNNVPLFEE305 pKa = 6.23LDD307 pKa = 3.67PNGFTYY313 pKa = 10.48HH314 pKa = 6.68FSKK317 pKa = 10.68LGNSTRR323 pKa = 11.84IFCVQRR329 pKa = 11.84DD330 pKa = 4.06LSYY333 pKa = 10.37MVLPAGQGLKK343 pKa = 10.5
Molecular weight: 38.53 kDa
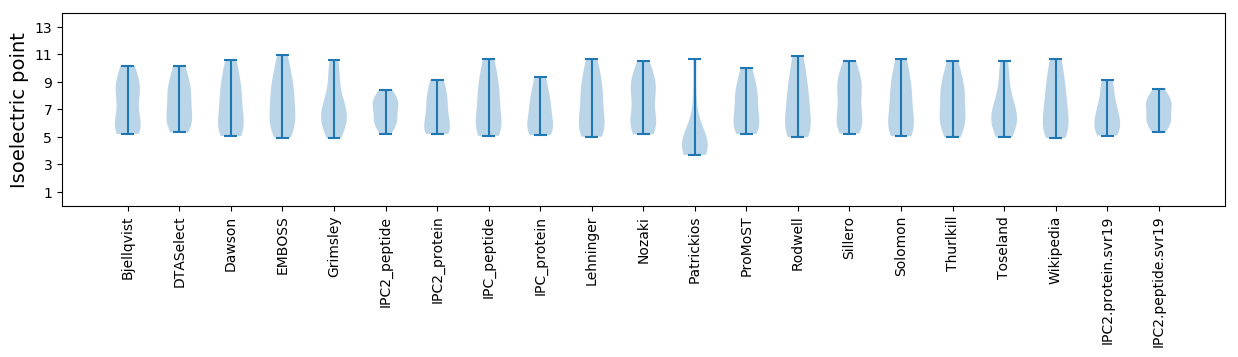
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5QM02|W5QM02_9REOV NS4 OS=Orungo virus OX=40058 PE=4 SV=1
MM1 pKa = 6.37MTSKK5 pKa = 10.74RR6 pKa = 11.84MNQWIRR12 pKa = 11.84EE13 pKa = 4.06AASRR17 pKa = 11.84VIEE20 pKa = 4.18SGRR23 pKa = 11.84KK24 pKa = 9.45LEE26 pKa = 3.99MPRR29 pKa = 11.84VNSLPIKK36 pKa = 9.89RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84LEE41 pKa = 3.67EE42 pKa = 3.66VRR44 pKa = 11.84IQSSGKK50 pKa = 9.86VEE52 pKa = 3.9MEE54 pKa = 3.55GDD56 pKa = 3.0KK57 pKa = 10.89LRR59 pKa = 11.84RR60 pKa = 11.84LKK62 pKa = 10.83AEE64 pKa = 3.87RR65 pKa = 11.84MEE67 pKa = 4.25LEE69 pKa = 3.67ILAFRR74 pKa = 11.84IKK76 pKa = 9.84TNNLIGALIQITRR89 pKa = 11.84WTEE92 pKa = 3.88KK93 pKa = 9.5IDD95 pKa = 3.84QEE97 pKa = 4.41IRR99 pKa = 11.84NLNMDD104 pKa = 4.44LQPNEE109 pKa = 4.15LQTPLSQSTTAVDD122 pKa = 4.36LRR124 pKa = 11.84GQDD127 pKa = 4.94RR128 pKa = 11.84MDD130 pKa = 3.38TSNN133 pKa = 3.3
MM1 pKa = 6.37MTSKK5 pKa = 10.74RR6 pKa = 11.84MNQWIRR12 pKa = 11.84EE13 pKa = 4.06AASRR17 pKa = 11.84VIEE20 pKa = 4.18SGRR23 pKa = 11.84KK24 pKa = 9.45LEE26 pKa = 3.99MPRR29 pKa = 11.84VNSLPIKK36 pKa = 9.89RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84LEE41 pKa = 3.67EE42 pKa = 3.66VRR44 pKa = 11.84IQSSGKK50 pKa = 9.86VEE52 pKa = 3.9MEE54 pKa = 3.55GDD56 pKa = 3.0KK57 pKa = 10.89LRR59 pKa = 11.84RR60 pKa = 11.84LKK62 pKa = 10.83AEE64 pKa = 3.87RR65 pKa = 11.84MEE67 pKa = 4.25LEE69 pKa = 3.67ILAFRR74 pKa = 11.84IKK76 pKa = 9.84TNNLIGALIQITRR89 pKa = 11.84WTEE92 pKa = 3.88KK93 pKa = 9.5IDD95 pKa = 3.84QEE97 pKa = 4.41IRR99 pKa = 11.84NLNMDD104 pKa = 4.44LQPNEE109 pKa = 4.15LQTPLSQSTTAVDD122 pKa = 4.36LRR124 pKa = 11.84GQDD127 pKa = 4.94RR128 pKa = 11.84MDD130 pKa = 3.38TSNN133 pKa = 3.3
Molecular weight: 15.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6209 |
133 |
1302 |
564.5 |
64.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.022 ± 0.621 | 0.902 ± 0.223 |
6.056 ± 0.303 | 6.684 ± 0.482 |
4.252 ± 0.521 | 6.265 ± 0.418 |
2.174 ± 0.339 | 6.507 ± 0.331 |
5.138 ± 0.559 | 9.019 ± 0.386 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.656 ± 0.258 | 3.769 ± 0.278 |
3.994 ± 0.379 | 3.64 ± 0.311 |
7.57 ± 0.426 | 5.991 ± 0.467 |
5.589 ± 0.278 | 7.103 ± 0.247 |
1.305 ± 0.213 | 3.366 ± 0.342 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
