
Nitrosomonas nitrosa
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosomonas
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
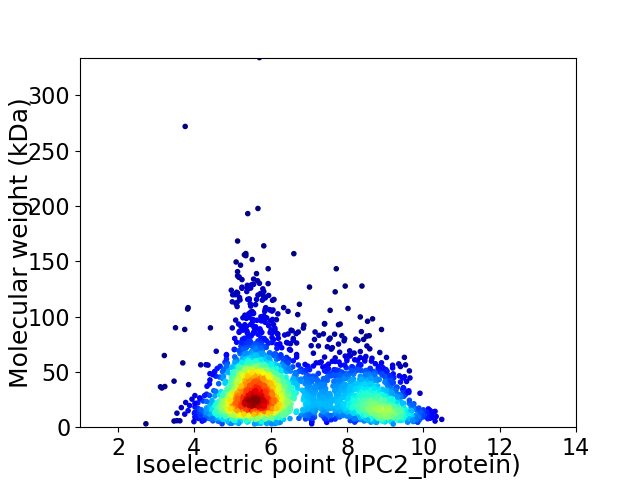
Virtual 2D-PAGE plot for 2883 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4UP56|A0A1I4UP56_9PROT Uncharacterized protein OS=Nitrosomonas nitrosa OX=52442 GN=SAMN05421880_1503 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.15KK3 pKa = 9.67IRR5 pKa = 11.84LICMIIALAGCLSAGPIWSADD26 pKa = 3.43GEE28 pKa = 4.64GGGDD32 pKa = 3.56FGGDD36 pKa = 3.25FDD38 pKa = 5.51GGGDD42 pKa = 3.57FGGDD46 pKa = 3.37FDD48 pKa = 5.75SSGDD52 pKa = 3.51FGGDD56 pKa = 2.93FDD58 pKa = 5.26YY59 pKa = 11.7GGDD62 pKa = 3.61FSGDD66 pKa = 3.14FDD68 pKa = 4.99SDD70 pKa = 3.79YY71 pKa = 11.69GGDD74 pKa = 3.45FGGDD78 pKa = 3.3FDD80 pKa = 4.93QGSEE84 pKa = 3.9FDD86 pKa = 3.36QGGEE90 pKa = 3.94FDD92 pKa = 3.45QGSDD96 pKa = 3.52FEE98 pKa = 4.84QDD100 pKa = 4.0FDD102 pKa = 3.72QQDD105 pKa = 3.49YY106 pKa = 10.69FGQEE110 pKa = 3.85EE111 pKa = 4.13FDD113 pKa = 3.67QDD115 pKa = 3.17GSEE118 pKa = 4.29YY119 pKa = 11.11GSVDD123 pKa = 3.21PNEE126 pKa = 5.7AFDD129 pKa = 3.41QDD131 pKa = 3.68NRR133 pKa = 11.84FDD135 pKa = 3.64QQQDD139 pKa = 3.39FEE141 pKa = 4.56FADD144 pKa = 4.86DD145 pKa = 3.98SDD147 pKa = 3.79QAGDD151 pKa = 3.86FDD153 pKa = 5.34QNDD156 pKa = 4.31DD157 pKa = 4.37FDD159 pKa = 3.95QADD162 pKa = 3.73NFDD165 pKa = 4.16PQNDD169 pKa = 3.51ASLEE173 pKa = 4.33SNADD177 pKa = 3.54PVIEE181 pKa = 4.21SAPEE185 pKa = 3.61EE186 pKa = 4.53VLTQDD191 pKa = 3.93EE192 pKa = 5.09DD193 pKa = 3.96PAQEE197 pKa = 4.17DD198 pKa = 3.76QSTGMSEE205 pKa = 3.79PAQSNEE211 pKa = 3.67FALDD215 pKa = 3.8NADD218 pKa = 3.68TDD220 pKa = 4.02NKK222 pKa = 10.01STEE225 pKa = 4.01TDD227 pKa = 3.19QPRR230 pKa = 11.84DD231 pKa = 3.65DD232 pKa = 4.86PVAQSAGEE240 pKa = 4.05PNPEE244 pKa = 4.58SIEE247 pKa = 4.0TDD249 pKa = 3.26QVDD252 pKa = 3.98TVSEE256 pKa = 4.39STQTLPSQQEE266 pKa = 3.82DD267 pKa = 3.34KK268 pKa = 11.17AAEE271 pKa = 4.07NDD273 pKa = 3.62GLAQTGDD280 pKa = 3.47SALPDD285 pKa = 3.16QSLQSDD291 pKa = 4.11EE292 pKa = 4.63SSQAEE297 pKa = 4.05PTAQGEE303 pKa = 4.47DD304 pKa = 3.31ASQPTQSAQAGSDD317 pKa = 3.28AAGNPATQNDD327 pKa = 3.76VTDD330 pKa = 4.1QNDD333 pKa = 3.52TSNQNINIQNVQTVQNNGHH352 pKa = 6.13DD353 pKa = 3.62HH354 pKa = 6.48PVGHH358 pKa = 6.44GHH360 pKa = 6.76GGWHH364 pKa = 5.73GHH366 pKa = 4.85HH367 pKa = 7.02HH368 pKa = 5.94GHH370 pKa = 6.15HH371 pKa = 6.32HH372 pKa = 6.06HH373 pKa = 7.08HH374 pKa = 6.59GFDD377 pKa = 2.91IGFGLGLGLGLGWSAFSPFFPYY399 pKa = 10.7APVVPFAGFSYY410 pKa = 10.08GWGGFAPYY418 pKa = 10.19SSFGIYY424 pKa = 10.06SGFGSFGGVGFYY436 pKa = 11.09SNYY439 pKa = 9.8VPFGGVSLYY448 pKa = 10.71SGYY451 pKa = 11.2SRR453 pKa = 11.84FRR455 pKa = 11.84SFNRR459 pKa = 11.84FGGYY463 pKa = 9.45YY464 pKa = 9.13PPLGGFPIVAAAPPILPVPVMIAPPRR490 pKa = 11.84KK491 pKa = 7.63PTYY494 pKa = 7.62IQKK497 pKa = 10.21RR498 pKa = 11.84SVSRR502 pKa = 11.84PPPAEE507 pKa = 3.51NSYY510 pKa = 9.27YY511 pKa = 9.7WHH513 pKa = 6.8YY514 pKa = 10.73CRR516 pKa = 11.84NPAGYY521 pKa = 9.35YY522 pKa = 9.71PYY524 pKa = 10.76VRR526 pKa = 11.84KK527 pKa = 10.21CPGGWIKK534 pKa = 10.68VPPQPSS540 pKa = 2.89
MM1 pKa = 7.42KK2 pKa = 10.15KK3 pKa = 9.67IRR5 pKa = 11.84LICMIIALAGCLSAGPIWSADD26 pKa = 3.43GEE28 pKa = 4.64GGGDD32 pKa = 3.56FGGDD36 pKa = 3.25FDD38 pKa = 5.51GGGDD42 pKa = 3.57FGGDD46 pKa = 3.37FDD48 pKa = 5.75SSGDD52 pKa = 3.51FGGDD56 pKa = 2.93FDD58 pKa = 5.26YY59 pKa = 11.7GGDD62 pKa = 3.61FSGDD66 pKa = 3.14FDD68 pKa = 4.99SDD70 pKa = 3.79YY71 pKa = 11.69GGDD74 pKa = 3.45FGGDD78 pKa = 3.3FDD80 pKa = 4.93QGSEE84 pKa = 3.9FDD86 pKa = 3.36QGGEE90 pKa = 3.94FDD92 pKa = 3.45QGSDD96 pKa = 3.52FEE98 pKa = 4.84QDD100 pKa = 4.0FDD102 pKa = 3.72QQDD105 pKa = 3.49YY106 pKa = 10.69FGQEE110 pKa = 3.85EE111 pKa = 4.13FDD113 pKa = 3.67QDD115 pKa = 3.17GSEE118 pKa = 4.29YY119 pKa = 11.11GSVDD123 pKa = 3.21PNEE126 pKa = 5.7AFDD129 pKa = 3.41QDD131 pKa = 3.68NRR133 pKa = 11.84FDD135 pKa = 3.64QQQDD139 pKa = 3.39FEE141 pKa = 4.56FADD144 pKa = 4.86DD145 pKa = 3.98SDD147 pKa = 3.79QAGDD151 pKa = 3.86FDD153 pKa = 5.34QNDD156 pKa = 4.31DD157 pKa = 4.37FDD159 pKa = 3.95QADD162 pKa = 3.73NFDD165 pKa = 4.16PQNDD169 pKa = 3.51ASLEE173 pKa = 4.33SNADD177 pKa = 3.54PVIEE181 pKa = 4.21SAPEE185 pKa = 3.61EE186 pKa = 4.53VLTQDD191 pKa = 3.93EE192 pKa = 5.09DD193 pKa = 3.96PAQEE197 pKa = 4.17DD198 pKa = 3.76QSTGMSEE205 pKa = 3.79PAQSNEE211 pKa = 3.67FALDD215 pKa = 3.8NADD218 pKa = 3.68TDD220 pKa = 4.02NKK222 pKa = 10.01STEE225 pKa = 4.01TDD227 pKa = 3.19QPRR230 pKa = 11.84DD231 pKa = 3.65DD232 pKa = 4.86PVAQSAGEE240 pKa = 4.05PNPEE244 pKa = 4.58SIEE247 pKa = 4.0TDD249 pKa = 3.26QVDD252 pKa = 3.98TVSEE256 pKa = 4.39STQTLPSQQEE266 pKa = 3.82DD267 pKa = 3.34KK268 pKa = 11.17AAEE271 pKa = 4.07NDD273 pKa = 3.62GLAQTGDD280 pKa = 3.47SALPDD285 pKa = 3.16QSLQSDD291 pKa = 4.11EE292 pKa = 4.63SSQAEE297 pKa = 4.05PTAQGEE303 pKa = 4.47DD304 pKa = 3.31ASQPTQSAQAGSDD317 pKa = 3.28AAGNPATQNDD327 pKa = 3.76VTDD330 pKa = 4.1QNDD333 pKa = 3.52TSNQNINIQNVQTVQNNGHH352 pKa = 6.13DD353 pKa = 3.62HH354 pKa = 6.48PVGHH358 pKa = 6.44GHH360 pKa = 6.76GGWHH364 pKa = 5.73GHH366 pKa = 4.85HH367 pKa = 7.02HH368 pKa = 5.94GHH370 pKa = 6.15HH371 pKa = 6.32HH372 pKa = 6.06HH373 pKa = 7.08HH374 pKa = 6.59GFDD377 pKa = 2.91IGFGLGLGLGLGWSAFSPFFPYY399 pKa = 10.7APVVPFAGFSYY410 pKa = 10.08GWGGFAPYY418 pKa = 10.19SSFGIYY424 pKa = 10.06SGFGSFGGVGFYY436 pKa = 11.09SNYY439 pKa = 9.8VPFGGVSLYY448 pKa = 10.71SGYY451 pKa = 11.2SRR453 pKa = 11.84FRR455 pKa = 11.84SFNRR459 pKa = 11.84FGGYY463 pKa = 9.45YY464 pKa = 9.13PPLGGFPIVAAAPPILPVPVMIAPPRR490 pKa = 11.84KK491 pKa = 7.63PTYY494 pKa = 7.62IQKK497 pKa = 10.21RR498 pKa = 11.84SVSRR502 pKa = 11.84PPPAEE507 pKa = 3.51NSYY510 pKa = 9.27YY511 pKa = 9.7WHH513 pKa = 6.8YY514 pKa = 10.73CRR516 pKa = 11.84NPAGYY521 pKa = 9.35YY522 pKa = 9.71PYY524 pKa = 10.76VRR526 pKa = 11.84KK527 pKa = 10.21CPGGWIKK534 pKa = 10.68VPPQPSS540 pKa = 2.89
Molecular weight: 58.09 kDa
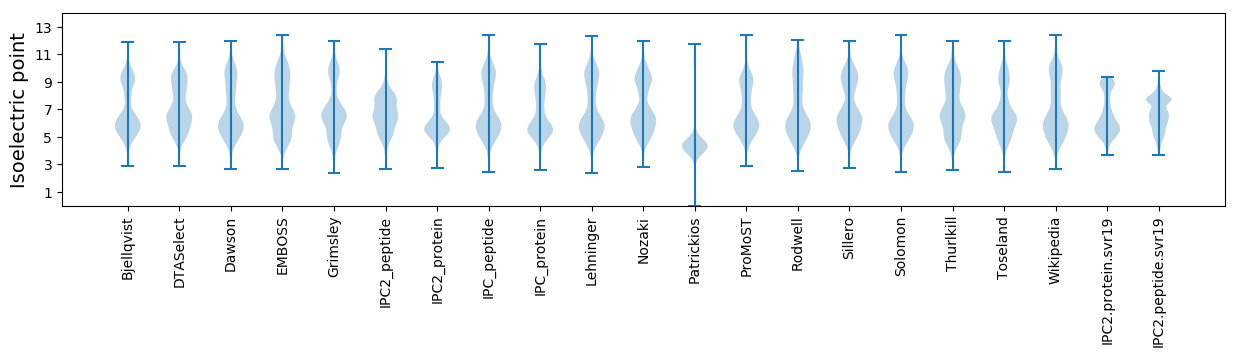
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4PZL8|A0A1I4PZL8_9PROT Uncharacterized protein OS=Nitrosomonas nitrosa OX=52442 GN=SAMN05421880_11328 PE=4 SV=1
MM1 pKa = 7.85IDD3 pKa = 3.63FTSALYY9 pKa = 10.62LGLHH13 pKa = 6.0HH14 pKa = 7.35PSRR17 pKa = 11.84LLQPWQQLTLGMPAALAEE35 pKa = 4.29PPGAAEE41 pKa = 4.32VAQKK45 pKa = 10.5LARR48 pKa = 11.84LQGCEE53 pKa = 4.22RR54 pKa = 11.84GVLMPSTLHH63 pKa = 6.74LFWDD67 pKa = 4.74LFGMLCRR74 pKa = 11.84QPVTLYY80 pKa = 10.85LDD82 pKa = 3.59EE83 pKa = 4.3GTYY86 pKa = 10.64AIARR90 pKa = 11.84WGMARR95 pKa = 11.84LSARR99 pKa = 11.84GVAIHH104 pKa = 6.76PFRR107 pKa = 11.84HH108 pKa = 6.05HH109 pKa = 7.36DD110 pKa = 3.83ANALLTKK117 pKa = 9.68ITRR120 pKa = 11.84QARR123 pKa = 11.84NRR125 pKa = 11.84KK126 pKa = 8.6YY127 pKa = 10.24PVVVANGFCPACGTAAPIGDD147 pKa = 3.87YY148 pKa = 11.01LDD150 pKa = 3.01IVRR153 pKa = 11.84RR154 pKa = 11.84FDD156 pKa = 3.44GLLVLDD162 pKa = 4.37DD163 pKa = 4.02TQALGVLGEE172 pKa = 4.26APALRR177 pKa = 11.84RR178 pKa = 11.84PYY180 pKa = 10.83GLGGGGILRR189 pKa = 11.84WSGHH193 pKa = 5.37TGPDD197 pKa = 2.98ILVGSSLAKK206 pKa = 10.46GLGVPVAVLTGSDD219 pKa = 2.91RR220 pKa = 11.84LLRR223 pKa = 11.84RR224 pKa = 11.84FTVASDD230 pKa = 3.49TRR232 pKa = 11.84VHH234 pKa = 6.92SSPPSLAVIQAAIHH248 pKa = 5.68ALAVNRR254 pKa = 11.84LQGNLLRR261 pKa = 11.84LRR263 pKa = 11.84LAQRR267 pKa = 11.84VGQFRR272 pKa = 11.84RR273 pKa = 11.84GLAGIGWSSTGGWFPVQTLCGITEE297 pKa = 3.98KK298 pKa = 10.97VAIRR302 pKa = 11.84LHH304 pKa = 5.55QRR306 pKa = 11.84LAQWGVQTVLTRR318 pKa = 11.84SRR320 pKa = 11.84HH321 pKa = 3.94NRR323 pKa = 11.84AARR326 pKa = 11.84LSFLLTARR334 pKa = 11.84HH335 pKa = 5.62RR336 pKa = 11.84AEE338 pKa = 5.74EE339 pKa = 3.98IDD341 pKa = 3.37QCIDD345 pKa = 3.33MLDD348 pKa = 3.41ALAPEE353 pKa = 4.92IEE355 pKa = 4.3APTNIEE361 pKa = 3.99APKK364 pKa = 10.04GRR366 pKa = 11.84PVLACQPRR374 pKa = 11.84GG375 pKa = 3.37
MM1 pKa = 7.85IDD3 pKa = 3.63FTSALYY9 pKa = 10.62LGLHH13 pKa = 6.0HH14 pKa = 7.35PSRR17 pKa = 11.84LLQPWQQLTLGMPAALAEE35 pKa = 4.29PPGAAEE41 pKa = 4.32VAQKK45 pKa = 10.5LARR48 pKa = 11.84LQGCEE53 pKa = 4.22RR54 pKa = 11.84GVLMPSTLHH63 pKa = 6.74LFWDD67 pKa = 4.74LFGMLCRR74 pKa = 11.84QPVTLYY80 pKa = 10.85LDD82 pKa = 3.59EE83 pKa = 4.3GTYY86 pKa = 10.64AIARR90 pKa = 11.84WGMARR95 pKa = 11.84LSARR99 pKa = 11.84GVAIHH104 pKa = 6.76PFRR107 pKa = 11.84HH108 pKa = 6.05HH109 pKa = 7.36DD110 pKa = 3.83ANALLTKK117 pKa = 9.68ITRR120 pKa = 11.84QARR123 pKa = 11.84NRR125 pKa = 11.84KK126 pKa = 8.6YY127 pKa = 10.24PVVVANGFCPACGTAAPIGDD147 pKa = 3.87YY148 pKa = 11.01LDD150 pKa = 3.01IVRR153 pKa = 11.84RR154 pKa = 11.84FDD156 pKa = 3.44GLLVLDD162 pKa = 4.37DD163 pKa = 4.02TQALGVLGEE172 pKa = 4.26APALRR177 pKa = 11.84RR178 pKa = 11.84PYY180 pKa = 10.83GLGGGGILRR189 pKa = 11.84WSGHH193 pKa = 5.37TGPDD197 pKa = 2.98ILVGSSLAKK206 pKa = 10.46GLGVPVAVLTGSDD219 pKa = 2.91RR220 pKa = 11.84LLRR223 pKa = 11.84RR224 pKa = 11.84FTVASDD230 pKa = 3.49TRR232 pKa = 11.84VHH234 pKa = 6.92SSPPSLAVIQAAIHH248 pKa = 5.68ALAVNRR254 pKa = 11.84LQGNLLRR261 pKa = 11.84LRR263 pKa = 11.84LAQRR267 pKa = 11.84VGQFRR272 pKa = 11.84RR273 pKa = 11.84GLAGIGWSSTGGWFPVQTLCGITEE297 pKa = 3.98KK298 pKa = 10.97VAIRR302 pKa = 11.84LHH304 pKa = 5.55QRR306 pKa = 11.84LAQWGVQTVLTRR318 pKa = 11.84SRR320 pKa = 11.84HH321 pKa = 3.94NRR323 pKa = 11.84AARR326 pKa = 11.84LSFLLTARR334 pKa = 11.84HH335 pKa = 5.62RR336 pKa = 11.84AEE338 pKa = 5.74EE339 pKa = 3.98IDD341 pKa = 3.37QCIDD345 pKa = 3.33MLDD348 pKa = 3.41ALAPEE353 pKa = 4.92IEE355 pKa = 4.3APTNIEE361 pKa = 3.99APKK364 pKa = 10.04GRR366 pKa = 11.84PVLACQPRR374 pKa = 11.84GG375 pKa = 3.37
Molecular weight: 40.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910118 |
26 |
2979 |
315.7 |
35.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.19 ± 0.045 | 1.003 ± 0.017 |
5.216 ± 0.038 | 6.069 ± 0.038 |
4.047 ± 0.033 | 6.918 ± 0.047 |
2.588 ± 0.025 | 6.804 ± 0.035 |
4.521 ± 0.037 | 10.817 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.022 | 3.739 ± 0.032 |
4.527 ± 0.03 | 4.28 ± 0.033 |
5.85 ± 0.04 | 5.934 ± 0.031 |
5.301 ± 0.035 | 6.515 ± 0.036 |
1.344 ± 0.019 | 2.906 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
