
Bovine foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Bovispumavirus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
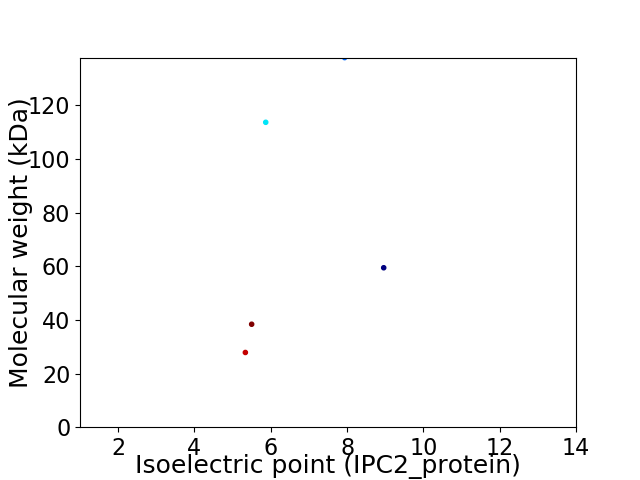
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9VDW6|J9VDW6_9RETR Tas OS=Bovine foamy virus OX=207343 GN=bel1 PE=4 SV=1
MM1 pKa = 7.7ASGGTPEE8 pKa = 4.3KK9 pKa = 10.96ARR11 pKa = 11.84VACRR15 pKa = 11.84RR16 pKa = 11.84VDD18 pKa = 3.69LSSFLAQPDD27 pKa = 4.52DD28 pKa = 4.08YY29 pKa = 10.64PTAADD34 pKa = 3.73SKK36 pKa = 11.1EE37 pKa = 3.96DD38 pKa = 4.39LILKK42 pKa = 7.48LACTTLFSEE51 pKa = 4.5KK52 pKa = 9.52HH53 pKa = 4.38AHH55 pKa = 6.78EE56 pKa = 4.5IYY58 pKa = 10.69EE59 pKa = 4.1NYY61 pKa = 9.97KK62 pKa = 10.36LHH64 pKa = 7.14LKK66 pKa = 10.13RR67 pKa = 11.84DD68 pKa = 3.63EE69 pKa = 4.08LRR71 pKa = 11.84GGKK74 pKa = 8.61EE75 pKa = 3.19WVIIYY80 pKa = 9.72SCKK83 pKa = 9.4HH84 pKa = 6.16CYY86 pKa = 7.86TVFMDD91 pKa = 4.06NSRR94 pKa = 11.84LTLDD98 pKa = 3.18PSGLFRR104 pKa = 11.84VIRR107 pKa = 11.84NKK109 pKa = 10.24KK110 pKa = 8.51GPYY113 pKa = 8.72MLCQMLTRR121 pKa = 11.84HH122 pKa = 5.4LTDD125 pKa = 2.96RR126 pKa = 11.84CNPRR130 pKa = 11.84TKK132 pKa = 10.14PFQSSSSLHH141 pKa = 6.27PNLVTEE147 pKa = 4.86DD148 pKa = 3.51PRR150 pKa = 11.84GAGGGTPGQHH160 pKa = 5.79TLGGDD165 pKa = 2.66QDD167 pKa = 3.8MRR169 pKa = 11.84VNTSGIKK176 pKa = 9.71PLSSLCQCARR186 pKa = 11.84DD187 pKa = 3.97DD188 pKa = 4.73PGRR191 pKa = 11.84SDD193 pKa = 4.31NPLEE197 pKa = 4.12MAEE200 pKa = 4.6PVQPWWTDD208 pKa = 2.79SSLEE212 pKa = 4.03PEE214 pKa = 3.72IATWVLGNPDD224 pKa = 2.66ATARR228 pKa = 11.84FWTGDD233 pKa = 3.4DD234 pKa = 4.47KK235 pKa = 11.78GPQEE239 pKa = 4.33WDD241 pKa = 3.27FDD243 pKa = 4.84DD244 pKa = 6.28DD245 pKa = 4.96LLGPP249 pKa = 4.64
MM1 pKa = 7.7ASGGTPEE8 pKa = 4.3KK9 pKa = 10.96ARR11 pKa = 11.84VACRR15 pKa = 11.84RR16 pKa = 11.84VDD18 pKa = 3.69LSSFLAQPDD27 pKa = 4.52DD28 pKa = 4.08YY29 pKa = 10.64PTAADD34 pKa = 3.73SKK36 pKa = 11.1EE37 pKa = 3.96DD38 pKa = 4.39LILKK42 pKa = 7.48LACTTLFSEE51 pKa = 4.5KK52 pKa = 9.52HH53 pKa = 4.38AHH55 pKa = 6.78EE56 pKa = 4.5IYY58 pKa = 10.69EE59 pKa = 4.1NYY61 pKa = 9.97KK62 pKa = 10.36LHH64 pKa = 7.14LKK66 pKa = 10.13RR67 pKa = 11.84DD68 pKa = 3.63EE69 pKa = 4.08LRR71 pKa = 11.84GGKK74 pKa = 8.61EE75 pKa = 3.19WVIIYY80 pKa = 9.72SCKK83 pKa = 9.4HH84 pKa = 6.16CYY86 pKa = 7.86TVFMDD91 pKa = 4.06NSRR94 pKa = 11.84LTLDD98 pKa = 3.18PSGLFRR104 pKa = 11.84VIRR107 pKa = 11.84NKK109 pKa = 10.24KK110 pKa = 8.51GPYY113 pKa = 8.72MLCQMLTRR121 pKa = 11.84HH122 pKa = 5.4LTDD125 pKa = 2.96RR126 pKa = 11.84CNPRR130 pKa = 11.84TKK132 pKa = 10.14PFQSSSSLHH141 pKa = 6.27PNLVTEE147 pKa = 4.86DD148 pKa = 3.51PRR150 pKa = 11.84GAGGGTPGQHH160 pKa = 5.79TLGGDD165 pKa = 2.66QDD167 pKa = 3.8MRR169 pKa = 11.84VNTSGIKK176 pKa = 9.71PLSSLCQCARR186 pKa = 11.84DD187 pKa = 3.97DD188 pKa = 4.73PGRR191 pKa = 11.84SDD193 pKa = 4.31NPLEE197 pKa = 4.12MAEE200 pKa = 4.6PVQPWWTDD208 pKa = 2.79SSLEE212 pKa = 4.03PEE214 pKa = 3.72IATWVLGNPDD224 pKa = 2.66ATARR228 pKa = 11.84FWTGDD233 pKa = 3.4DD234 pKa = 4.47KK235 pKa = 11.78GPQEE239 pKa = 4.33WDD241 pKa = 3.27FDD243 pKa = 4.84DD244 pKa = 6.28DD245 pKa = 4.96LLGPP249 pKa = 4.64
Molecular weight: 27.87 kDa
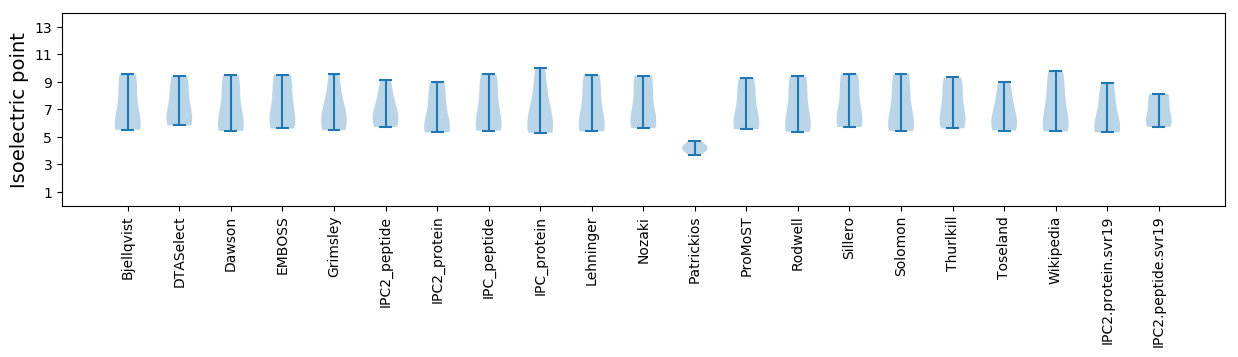
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9V9W7|J9V9W7_9RETR Bet OS=Bovine foamy virus OX=207343 GN=bel2 PE=4 SV=1
MM1 pKa = 7.76ALNDD5 pKa = 4.11FDD7 pKa = 6.17PIALQGYY14 pKa = 8.63LPAPRR19 pKa = 11.84VLQHH23 pKa = 6.42NDD25 pKa = 2.94IIICRR30 pKa = 11.84ATSGPWGIGDD40 pKa = 4.22RR41 pKa = 11.84YY42 pKa = 10.43NLIRR46 pKa = 11.84IHH48 pKa = 6.27LQDD51 pKa = 4.15PAGQPLPIPQWEE63 pKa = 4.51PIPNRR68 pKa = 11.84TANPRR73 pKa = 11.84TQPYY77 pKa = 8.94PVVSAPMATLEE88 pKa = 4.33NILNNFHH95 pKa = 6.77IPHH98 pKa = 6.13GVSRR102 pKa = 11.84YY103 pKa = 9.83GPLEE107 pKa = 4.3GGDD110 pKa = 3.88YY111 pKa = 10.88QPGEE115 pKa = 4.16QYY117 pKa = 11.05SQGFCPVTQAEE128 pKa = 4.26IALLNGQHH136 pKa = 6.51LEE138 pKa = 4.08EE139 pKa = 5.54EE140 pKa = 4.21ITILRR145 pKa = 11.84EE146 pKa = 3.5ITHH149 pKa = 7.18RR150 pKa = 11.84LMQGVRR156 pKa = 11.84PPAAPQGPAPPPPPAQPPAPLPAPPVGPPPPAVPAPAPGPMPVPQHH202 pKa = 6.46LPITHH207 pKa = 6.16IRR209 pKa = 11.84AVIGEE214 pKa = 4.3TPANIRR220 pKa = 11.84EE221 pKa = 4.21VPLWLARR228 pKa = 11.84AVPALQGVYY237 pKa = 9.48PVQDD241 pKa = 2.69AVMRR245 pKa = 11.84SRR247 pKa = 11.84TVNALTVRR255 pKa = 11.84HH256 pKa = 6.6PGLALEE262 pKa = 4.36PLEE265 pKa = 4.46CGSWQEE271 pKa = 4.11CLAALWQRR279 pKa = 11.84TFGATALHH287 pKa = 6.59ALGDD291 pKa = 4.0TLGQIANSDD300 pKa = 4.32GIVMAIEE307 pKa = 4.45LGLLFSDD314 pKa = 5.57DD315 pKa = 3.29NWDD318 pKa = 4.02LVWGICRR325 pKa = 11.84RR326 pKa = 11.84FLPGQAVCVAVQARR340 pKa = 11.84LDD342 pKa = 4.08PLPDD346 pKa = 3.22NATRR350 pKa = 11.84IVMISHH356 pKa = 8.19IIRR359 pKa = 11.84DD360 pKa = 3.64VYY362 pKa = 10.98AILGLDD368 pKa = 3.36PLGRR372 pKa = 11.84PMQQTLPRR380 pKa = 11.84RR381 pKa = 11.84NNQPPRR387 pKa = 11.84QQPQRR392 pKa = 11.84RR393 pKa = 11.84QQPRR397 pKa = 11.84RR398 pKa = 11.84TGNQEE403 pKa = 3.16EE404 pKa = 4.05RR405 pKa = 11.84GQRR408 pKa = 11.84NRR410 pKa = 11.84GRR412 pKa = 11.84QNAQTPRR419 pKa = 11.84QEE421 pKa = 4.19GNRR424 pKa = 11.84LQNSQLPGPQDD435 pKa = 3.44YY436 pKa = 10.0PNNSNQPRR444 pKa = 11.84YY445 pKa = 8.98PLRR448 pKa = 11.84PNPQQPQRR456 pKa = 11.84YY457 pKa = 8.09GQEE460 pKa = 3.91QNRR463 pKa = 11.84GNNPNPYY470 pKa = 9.58RR471 pKa = 11.84QPTPGNGNQNRR482 pKa = 11.84NFSRR486 pKa = 11.84GPAPVNEE493 pKa = 4.26QSRR496 pKa = 11.84GRR498 pKa = 11.84GRR500 pKa = 11.84NSQGTNNTGSSAVHH514 pKa = 5.3SVRR517 pKa = 11.84LTSAAPPIPPQDD529 pKa = 3.61AGTPPTPSGNQGQSSS544 pKa = 3.44
MM1 pKa = 7.76ALNDD5 pKa = 4.11FDD7 pKa = 6.17PIALQGYY14 pKa = 8.63LPAPRR19 pKa = 11.84VLQHH23 pKa = 6.42NDD25 pKa = 2.94IIICRR30 pKa = 11.84ATSGPWGIGDD40 pKa = 4.22RR41 pKa = 11.84YY42 pKa = 10.43NLIRR46 pKa = 11.84IHH48 pKa = 6.27LQDD51 pKa = 4.15PAGQPLPIPQWEE63 pKa = 4.51PIPNRR68 pKa = 11.84TANPRR73 pKa = 11.84TQPYY77 pKa = 8.94PVVSAPMATLEE88 pKa = 4.33NILNNFHH95 pKa = 6.77IPHH98 pKa = 6.13GVSRR102 pKa = 11.84YY103 pKa = 9.83GPLEE107 pKa = 4.3GGDD110 pKa = 3.88YY111 pKa = 10.88QPGEE115 pKa = 4.16QYY117 pKa = 11.05SQGFCPVTQAEE128 pKa = 4.26IALLNGQHH136 pKa = 6.51LEE138 pKa = 4.08EE139 pKa = 5.54EE140 pKa = 4.21ITILRR145 pKa = 11.84EE146 pKa = 3.5ITHH149 pKa = 7.18RR150 pKa = 11.84LMQGVRR156 pKa = 11.84PPAAPQGPAPPPPPAQPPAPLPAPPVGPPPPAVPAPAPGPMPVPQHH202 pKa = 6.46LPITHH207 pKa = 6.16IRR209 pKa = 11.84AVIGEE214 pKa = 4.3TPANIRR220 pKa = 11.84EE221 pKa = 4.21VPLWLARR228 pKa = 11.84AVPALQGVYY237 pKa = 9.48PVQDD241 pKa = 2.69AVMRR245 pKa = 11.84SRR247 pKa = 11.84TVNALTVRR255 pKa = 11.84HH256 pKa = 6.6PGLALEE262 pKa = 4.36PLEE265 pKa = 4.46CGSWQEE271 pKa = 4.11CLAALWQRR279 pKa = 11.84TFGATALHH287 pKa = 6.59ALGDD291 pKa = 4.0TLGQIANSDD300 pKa = 4.32GIVMAIEE307 pKa = 4.45LGLLFSDD314 pKa = 5.57DD315 pKa = 3.29NWDD318 pKa = 4.02LVWGICRR325 pKa = 11.84RR326 pKa = 11.84FLPGQAVCVAVQARR340 pKa = 11.84LDD342 pKa = 4.08PLPDD346 pKa = 3.22NATRR350 pKa = 11.84IVMISHH356 pKa = 8.19IIRR359 pKa = 11.84DD360 pKa = 3.64VYY362 pKa = 10.98AILGLDD368 pKa = 3.36PLGRR372 pKa = 11.84PMQQTLPRR380 pKa = 11.84RR381 pKa = 11.84NNQPPRR387 pKa = 11.84QQPQRR392 pKa = 11.84RR393 pKa = 11.84QQPRR397 pKa = 11.84RR398 pKa = 11.84TGNQEE403 pKa = 3.16EE404 pKa = 4.05RR405 pKa = 11.84GQRR408 pKa = 11.84NRR410 pKa = 11.84GRR412 pKa = 11.84QNAQTPRR419 pKa = 11.84QEE421 pKa = 4.19GNRR424 pKa = 11.84LQNSQLPGPQDD435 pKa = 3.44YY436 pKa = 10.0PNNSNQPRR444 pKa = 11.84YY445 pKa = 8.98PLRR448 pKa = 11.84PNPQQPQRR456 pKa = 11.84YY457 pKa = 8.09GQEE460 pKa = 3.91QNRR463 pKa = 11.84GNNPNPYY470 pKa = 9.58RR471 pKa = 11.84QPTPGNGNQNRR482 pKa = 11.84NFSRR486 pKa = 11.84GPAPVNEE493 pKa = 4.26QSRR496 pKa = 11.84GRR498 pKa = 11.84GRR500 pKa = 11.84NSQGTNNTGSSAVHH514 pKa = 5.3SVRR517 pKa = 11.84LTSAAPPIPPQDD529 pKa = 3.61AGTPPTPSGNQGQSSS544 pKa = 3.44
Molecular weight: 59.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3338 |
249 |
1220 |
667.6 |
75.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.201 ± 0.746 | 2.067 ± 0.279 |
5.153 ± 0.497 | 5.362 ± 0.523 |
2.337 ± 0.283 | 5.992 ± 0.781 |
2.966 ± 0.23 | 5.752 ± 0.528 |
4.823 ± 1.258 | 9.976 ± 0.882 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.588 ± 0.278 | 4.643 ± 0.515 |
8.328 ± 1.78 | 5.812 ± 0.838 |
5.692 ± 0.834 | 5.932 ± 0.691 |
6.171 ± 0.489 | 5.213 ± 0.436 |
2.397 ± 0.331 | 3.595 ± 0.519 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
