
Sewage-associated circular DNA virus-17
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
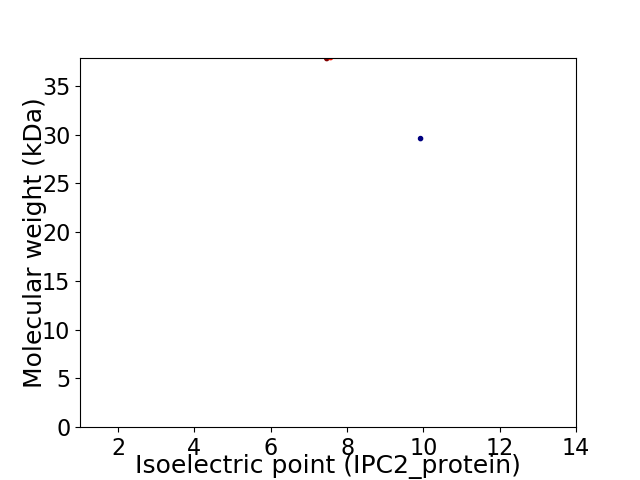
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGL4|A0A0B4UGL4_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-17 OX=1592084 PE=4 SV=1
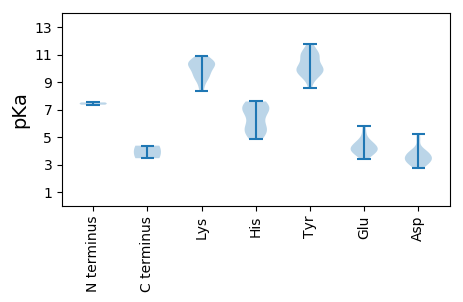
MM1 pKa = 7.52TPTPKK6 pKa = 10.13KK7 pKa = 10.44SYY9 pKa = 10.84CFTLNNYY16 pKa = 8.54TEE18 pKa = 5.12DD19 pKa = 3.29EE20 pKa = 4.15LAAIRR25 pKa = 11.84GVCSTLAQYY34 pKa = 11.0AIVGRR39 pKa = 11.84EE40 pKa = 3.8VGEE43 pKa = 3.74QGTRR47 pKa = 11.84HH48 pKa = 4.85LQGYY52 pKa = 9.51IRR54 pKa = 11.84FTRR57 pKa = 11.84AYY59 pKa = 9.56RR60 pKa = 11.84FPTIKK65 pKa = 10.27DD66 pKa = 3.12RR67 pKa = 11.84YY68 pKa = 9.88LPRR71 pKa = 11.84CHH73 pKa = 7.59IEE75 pKa = 3.81VSRR78 pKa = 11.84GSPRR82 pKa = 11.84QNRR85 pKa = 11.84DD86 pKa = 2.93YY87 pKa = 10.87CSKK90 pKa = 10.9DD91 pKa = 3.31GEE93 pKa = 4.01FDD95 pKa = 3.45EE96 pKa = 5.47YY97 pKa = 11.79GEE99 pKa = 4.33LPEE102 pKa = 5.82DD103 pKa = 3.63RR104 pKa = 11.84ASSSRR109 pKa = 11.84EE110 pKa = 3.43DD111 pKa = 3.5LARR114 pKa = 11.84EE115 pKa = 4.38FARR118 pKa = 11.84DD119 pKa = 3.46ATDD122 pKa = 2.78GRR124 pKa = 11.84GGVRR128 pKa = 11.84RR129 pKa = 11.84FAEE132 pKa = 4.61SNPGTYY138 pKa = 10.07YY139 pKa = 10.86FSGHH143 pKa = 5.65NLLRR147 pKa = 11.84NHH149 pKa = 7.26WALQAPTARR158 pKa = 11.84PGITVDD164 pKa = 3.22WMHH167 pKa = 6.83GLPGVGKK174 pKa = 9.61SRR176 pKa = 11.84RR177 pKa = 11.84AHH179 pKa = 5.87EE180 pKa = 4.65RR181 pKa = 11.84LPDD184 pKa = 3.85AYY186 pKa = 9.91IKK188 pKa = 10.49DD189 pKa = 4.08PRR191 pKa = 11.84TKK193 pKa = 8.56WWNGYY198 pKa = 9.2FGEE201 pKa = 4.6KK202 pKa = 9.89EE203 pKa = 4.18VIIDD207 pKa = 3.68DD208 pKa = 4.93FGPQGIDD215 pKa = 2.87INHH218 pKa = 7.3LLRR221 pKa = 11.84WFDD224 pKa = 3.52RR225 pKa = 11.84YY226 pKa = 10.96KK227 pKa = 10.58CTVEE231 pKa = 4.05YY232 pKa = 10.09KK233 pKa = 10.64GGMMPLMAEE242 pKa = 4.19TFIVTSNYY250 pKa = 9.37HH251 pKa = 5.4PRR253 pKa = 11.84EE254 pKa = 3.86VFQTGRR260 pKa = 11.84STDD263 pKa = 3.48QQDD266 pKa = 3.07NTMYY270 pKa = 10.95DD271 pKa = 3.71HH272 pKa = 7.14PQLPALMRR280 pKa = 11.84RR281 pKa = 11.84INIIHH286 pKa = 5.5MQYY289 pKa = 9.7IIEE292 pKa = 4.38YY293 pKa = 9.97ISRR296 pKa = 11.84CMDD299 pKa = 3.44ACNEE303 pKa = 3.52ASEE306 pKa = 4.3IVKK309 pKa = 10.17EE310 pKa = 4.08IIKK313 pKa = 10.34KK314 pKa = 9.41IMRR317 pKa = 11.84RR318 pKa = 11.84VPAPGAEE325 pKa = 4.0IAA327 pKa = 4.37
MM1 pKa = 7.52TPTPKK6 pKa = 10.13KK7 pKa = 10.44SYY9 pKa = 10.84CFTLNNYY16 pKa = 8.54TEE18 pKa = 5.12DD19 pKa = 3.29EE20 pKa = 4.15LAAIRR25 pKa = 11.84GVCSTLAQYY34 pKa = 11.0AIVGRR39 pKa = 11.84EE40 pKa = 3.8VGEE43 pKa = 3.74QGTRR47 pKa = 11.84HH48 pKa = 4.85LQGYY52 pKa = 9.51IRR54 pKa = 11.84FTRR57 pKa = 11.84AYY59 pKa = 9.56RR60 pKa = 11.84FPTIKK65 pKa = 10.27DD66 pKa = 3.12RR67 pKa = 11.84YY68 pKa = 9.88LPRR71 pKa = 11.84CHH73 pKa = 7.59IEE75 pKa = 3.81VSRR78 pKa = 11.84GSPRR82 pKa = 11.84QNRR85 pKa = 11.84DD86 pKa = 2.93YY87 pKa = 10.87CSKK90 pKa = 10.9DD91 pKa = 3.31GEE93 pKa = 4.01FDD95 pKa = 3.45EE96 pKa = 5.47YY97 pKa = 11.79GEE99 pKa = 4.33LPEE102 pKa = 5.82DD103 pKa = 3.63RR104 pKa = 11.84ASSSRR109 pKa = 11.84EE110 pKa = 3.43DD111 pKa = 3.5LARR114 pKa = 11.84EE115 pKa = 4.38FARR118 pKa = 11.84DD119 pKa = 3.46ATDD122 pKa = 2.78GRR124 pKa = 11.84GGVRR128 pKa = 11.84RR129 pKa = 11.84FAEE132 pKa = 4.61SNPGTYY138 pKa = 10.07YY139 pKa = 10.86FSGHH143 pKa = 5.65NLLRR147 pKa = 11.84NHH149 pKa = 7.26WALQAPTARR158 pKa = 11.84PGITVDD164 pKa = 3.22WMHH167 pKa = 6.83GLPGVGKK174 pKa = 9.61SRR176 pKa = 11.84RR177 pKa = 11.84AHH179 pKa = 5.87EE180 pKa = 4.65RR181 pKa = 11.84LPDD184 pKa = 3.85AYY186 pKa = 9.91IKK188 pKa = 10.49DD189 pKa = 4.08PRR191 pKa = 11.84TKK193 pKa = 8.56WWNGYY198 pKa = 9.2FGEE201 pKa = 4.6KK202 pKa = 9.89EE203 pKa = 4.18VIIDD207 pKa = 3.68DD208 pKa = 4.93FGPQGIDD215 pKa = 2.87INHH218 pKa = 7.3LLRR221 pKa = 11.84WFDD224 pKa = 3.52RR225 pKa = 11.84YY226 pKa = 10.96KK227 pKa = 10.58CTVEE231 pKa = 4.05YY232 pKa = 10.09KK233 pKa = 10.64GGMMPLMAEE242 pKa = 4.19TFIVTSNYY250 pKa = 9.37HH251 pKa = 5.4PRR253 pKa = 11.84EE254 pKa = 3.86VFQTGRR260 pKa = 11.84STDD263 pKa = 3.48QQDD266 pKa = 3.07NTMYY270 pKa = 10.95DD271 pKa = 3.71HH272 pKa = 7.14PQLPALMRR280 pKa = 11.84RR281 pKa = 11.84INIIHH286 pKa = 5.5MQYY289 pKa = 9.7IIEE292 pKa = 4.38YY293 pKa = 9.97ISRR296 pKa = 11.84CMDD299 pKa = 3.44ACNEE303 pKa = 3.52ASEE306 pKa = 4.3IVKK309 pKa = 10.17EE310 pKa = 4.08IIKK313 pKa = 10.34KK314 pKa = 9.41IMRR317 pKa = 11.84RR318 pKa = 11.84VPAPGAEE325 pKa = 4.0IAA327 pKa = 4.37
Molecular weight: 37.81 kDa
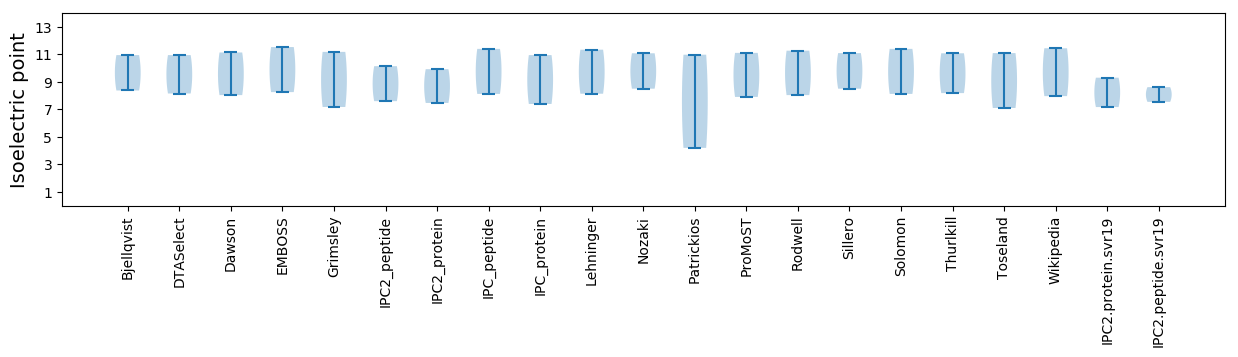
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGL4|A0A0B4UGL4_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-17 OX=1592084 PE=4 SV=1
MM1 pKa = 7.35AFKK4 pKa = 10.42RR5 pKa = 11.84KK6 pKa = 9.77RR7 pKa = 11.84IMARR11 pKa = 11.84RR12 pKa = 11.84SGFKK16 pKa = 9.8KK17 pKa = 10.05RR18 pKa = 11.84RR19 pKa = 11.84ISRR22 pKa = 11.84KK23 pKa = 9.26SRR25 pKa = 11.84FARR28 pKa = 11.84KK29 pKa = 9.37KK30 pKa = 9.01RR31 pKa = 11.84GSGTRR36 pKa = 11.84SWTQGGSRR44 pKa = 11.84AMDD47 pKa = 3.04TRR49 pKa = 11.84FRR51 pKa = 11.84SRR53 pKa = 11.84KK54 pKa = 9.13LLNSGWRR61 pKa = 11.84RR62 pKa = 11.84HH63 pKa = 5.39LWGQTLHH70 pKa = 6.98LSPSRR75 pKa = 11.84SALPVPIVQTSGTILGKK92 pKa = 10.8GSAVLYY98 pKa = 9.98WPTFNNAAPSAGNAFWTTAGGLNEE122 pKa = 4.64LDD124 pKa = 4.11AGVTAPTFTSEE135 pKa = 3.71NLIIRR140 pKa = 11.84GGRR143 pKa = 11.84IGITLTQLGSVSDD156 pKa = 4.71DD157 pKa = 2.95IGITVYY163 pKa = 10.08TLAMAKK169 pKa = 8.33NTATGRR175 pKa = 11.84LVTPVPWGVSIDD187 pKa = 3.84SAPDD191 pKa = 3.26FAEE194 pKa = 4.2LGKK197 pKa = 10.42ILDD200 pKa = 3.9RR201 pKa = 11.84KK202 pKa = 10.4DD203 pKa = 3.98YY204 pKa = 11.45IMDD207 pKa = 4.0ANFRR211 pKa = 11.84AVTIEE216 pKa = 4.83RR217 pKa = 11.84RR218 pKa = 11.84LKK220 pKa = 9.51CQKK223 pKa = 10.21IDD225 pKa = 3.29MDD227 pKa = 3.7QYY229 pKa = 11.19SIQGGQQIAYY239 pKa = 9.74CVVTTNLTSTTNVTTNFLIYY259 pKa = 10.21HH260 pKa = 6.89DD261 pKa = 5.25LSFSNTAA268 pKa = 3.46
MM1 pKa = 7.35AFKK4 pKa = 10.42RR5 pKa = 11.84KK6 pKa = 9.77RR7 pKa = 11.84IMARR11 pKa = 11.84RR12 pKa = 11.84SGFKK16 pKa = 9.8KK17 pKa = 10.05RR18 pKa = 11.84RR19 pKa = 11.84ISRR22 pKa = 11.84KK23 pKa = 9.26SRR25 pKa = 11.84FARR28 pKa = 11.84KK29 pKa = 9.37KK30 pKa = 9.01RR31 pKa = 11.84GSGTRR36 pKa = 11.84SWTQGGSRR44 pKa = 11.84AMDD47 pKa = 3.04TRR49 pKa = 11.84FRR51 pKa = 11.84SRR53 pKa = 11.84KK54 pKa = 9.13LLNSGWRR61 pKa = 11.84RR62 pKa = 11.84HH63 pKa = 5.39LWGQTLHH70 pKa = 6.98LSPSRR75 pKa = 11.84SALPVPIVQTSGTILGKK92 pKa = 10.8GSAVLYY98 pKa = 9.98WPTFNNAAPSAGNAFWTTAGGLNEE122 pKa = 4.64LDD124 pKa = 4.11AGVTAPTFTSEE135 pKa = 3.71NLIIRR140 pKa = 11.84GGRR143 pKa = 11.84IGITLTQLGSVSDD156 pKa = 4.71DD157 pKa = 2.95IGITVYY163 pKa = 10.08TLAMAKK169 pKa = 8.33NTATGRR175 pKa = 11.84LVTPVPWGVSIDD187 pKa = 3.84SAPDD191 pKa = 3.26FAEE194 pKa = 4.2LGKK197 pKa = 10.42ILDD200 pKa = 3.9RR201 pKa = 11.84KK202 pKa = 10.4DD203 pKa = 3.98YY204 pKa = 11.45IMDD207 pKa = 4.0ANFRR211 pKa = 11.84AVTIEE216 pKa = 4.83RR217 pKa = 11.84RR218 pKa = 11.84LKK220 pKa = 9.51CQKK223 pKa = 10.21IDD225 pKa = 3.29MDD227 pKa = 3.7QYY229 pKa = 11.19SIQGGQQIAYY239 pKa = 9.74CVVTTNLTSTTNVTTNFLIYY259 pKa = 10.21HH260 pKa = 6.89DD261 pKa = 5.25LSFSNTAA268 pKa = 3.46
Molecular weight: 29.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
595 |
268 |
327 |
297.5 |
33.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.395 ± 0.459 | 1.513 ± 0.432 |
5.378 ± 0.508 | 4.538 ± 1.718 |
3.866 ± 0.135 | 8.235 ± 0.406 |
2.185 ± 0.601 | 6.891 ± 0.098 |
4.538 ± 0.387 | 6.387 ± 0.817 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.689 ± 0.254 | 4.034 ± 0.25 |
4.706 ± 0.76 | 3.361 ± 0.002 |
9.748 ± 0.237 | 6.387 ± 1.238 |
7.899 ± 1.438 | 4.37 ± 0.271 |
1.849 ± 0.22 | 4.034 ± 1.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
