
Wenzhou tombus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.15
Get precalculated fractions of proteins
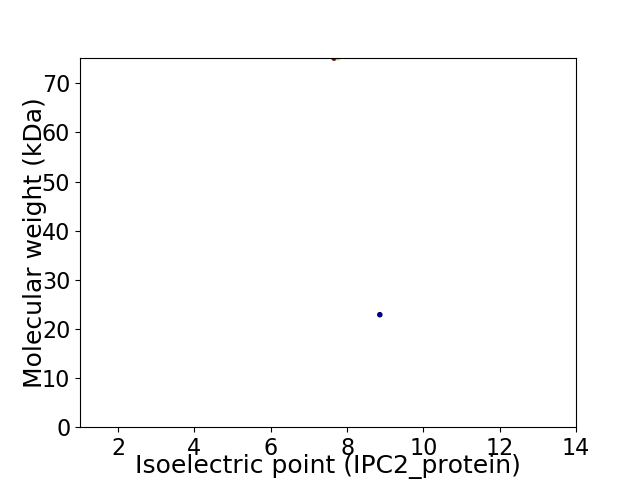
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH37|A0A1L3KH37_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 1 OX=1923662 PE=3 SV=1
MM1 pKa = 7.12VSAAFTFGCDD11 pKa = 2.81VGTSILLLVAVVGVVLGVRR30 pKa = 11.84WLRR33 pKa = 11.84SSTTGATLVAEE44 pKa = 4.51VDD46 pKa = 4.67LGPDD50 pKa = 3.06WTSVGVTTPYY60 pKa = 10.36QRR62 pKa = 11.84QLAYY66 pKa = 10.24QFKK69 pKa = 11.11AEE71 pKa = 4.13FGEE74 pKa = 4.25LRR76 pKa = 11.84YY77 pKa = 10.71NKK79 pKa = 10.0ANRR82 pKa = 11.84IIAGDD87 pKa = 3.6WCRR90 pKa = 11.84KK91 pKa = 9.38AMADD95 pKa = 3.15NDD97 pKa = 3.51VRR99 pKa = 11.84LTDD102 pKa = 3.73RR103 pKa = 11.84VRR105 pKa = 11.84LLPLAVEE112 pKa = 4.7LCLLPTRR119 pKa = 11.84HH120 pKa = 5.83AVAAAEE126 pKa = 3.89LAATHH131 pKa = 5.78EE132 pKa = 4.41VRR134 pKa = 11.84HH135 pKa = 5.7RR136 pKa = 11.84RR137 pKa = 11.84AAADD141 pKa = 3.81LPKK144 pKa = 9.71XGCPAILPGVTTRR157 pKa = 11.84VDD159 pKa = 3.51RR160 pKa = 11.84VGEE163 pKa = 3.76ACLRR167 pKa = 11.84VRR169 pKa = 11.84EE170 pKa = 4.23CVGVGVRR177 pKa = 11.84SGVDD181 pKa = 2.86RR182 pKa = 11.84SVRR185 pKa = 11.84YY186 pKa = 7.99MAGFGTGVRR195 pKa = 11.84YY196 pKa = 9.18GVHH199 pKa = 6.34CANLKK204 pKa = 9.66NLARR208 pKa = 11.84GIVEE212 pKa = 4.08RR213 pKa = 11.84VFHH216 pKa = 5.21VVRR219 pKa = 11.84DD220 pKa = 3.9GRR222 pKa = 11.84LAQAPQPRR230 pKa = 11.84FGVFSRR236 pKa = 11.84LNSVRR241 pKa = 11.84QRR243 pKa = 11.84LLAVTRR249 pKa = 11.84PTPVVARR256 pKa = 11.84EE257 pKa = 4.52DD258 pKa = 3.89YY259 pKa = 10.25PGLYY263 pKa = 9.31TGRR266 pKa = 11.84KK267 pKa = 7.19RR268 pKa = 11.84GIYY271 pKa = 9.33EE272 pKa = 3.9RR273 pKa = 11.84ALEE276 pKa = 4.21SLTVRR281 pKa = 11.84AINSRR286 pKa = 11.84DD287 pKa = 2.83AWVNTFVKK295 pKa = 10.53AEE297 pKa = 4.02KK298 pKa = 10.82VNLDD302 pKa = 3.82SKK304 pKa = 11.25GDD306 pKa = 3.66PAPRR310 pKa = 11.84VIQPRR315 pKa = 11.84SPRR318 pKa = 11.84YY319 pKa = 7.87NLEE322 pKa = 3.47VGRR325 pKa = 11.84YY326 pKa = 7.88LKK328 pKa = 10.18MFEE331 pKa = 5.17RR332 pKa = 11.84EE333 pKa = 3.93LCHH336 pKa = 6.7GFEE339 pKa = 4.6RR340 pKa = 11.84VWGYY344 pKa = 9.87PVVLKK349 pKa = 10.79GMNAQQVGGWMAAHH363 pKa = 7.16WGAFRR368 pKa = 11.84RR369 pKa = 11.84PVAVGLDD376 pKa = 3.23ASRR379 pKa = 11.84FDD381 pKa = 3.33QHH383 pKa = 7.36VSYY386 pKa = 11.07DD387 pKa = 3.46ALRR390 pKa = 11.84WEE392 pKa = 4.12HH393 pKa = 6.33SVYY396 pKa = 11.01NSTFHH401 pKa = 7.78SPEE404 pKa = 3.71LAKK407 pKa = 10.7LLRR410 pKa = 11.84WQLHH414 pKa = 4.33NRR416 pKa = 11.84GVARR420 pKa = 11.84TEE422 pKa = 4.41GYY424 pKa = 10.43RR425 pKa = 11.84IDD427 pKa = 3.8YY428 pKa = 9.84DD429 pKa = 3.33IRR431 pKa = 11.84GCRR434 pKa = 11.84MSGDD438 pKa = 3.53INTGMGNCLLMSSMVIAYY456 pKa = 9.09CEE458 pKa = 4.22SVGIEE463 pKa = 3.61YY464 pKa = 10.82RR465 pKa = 11.84LANNGDD471 pKa = 3.67DD472 pKa = 3.37CVLFVEE478 pKa = 4.57QQDD481 pKa = 4.05LVKK484 pKa = 10.93LSGIDD489 pKa = 3.07EE490 pKa = 4.19WMLDD494 pKa = 3.49FGFTLTRR501 pKa = 11.84EE502 pKa = 3.91EE503 pKa = 3.77PAYY506 pKa = 10.54VLEE509 pKa = 4.59HH510 pKa = 7.73VEE512 pKa = 4.54FCQARR517 pKa = 11.84PVFTSTGWRR526 pKa = 11.84MVRR529 pKa = 11.84DD530 pKa = 3.53PRR532 pKa = 11.84VAMSKK537 pKa = 10.67DD538 pKa = 3.69CVSLLGWEE546 pKa = 4.41SDD548 pKa = 3.87LDD550 pKa = 3.79LQYY553 pKa = 10.15WCHH556 pKa = 7.41AIGTCGASLTAGVPVWHH573 pKa = 6.47SWYY576 pKa = 10.3HH577 pKa = 5.22RR578 pKa = 11.84LQRR581 pKa = 11.84MGAVAPSGVVDD592 pKa = 3.57MVYY595 pKa = 10.95DD596 pKa = 3.56SGLGYY601 pKa = 9.75MSRR604 pKa = 11.84GVVGGEE610 pKa = 3.8VCAEE614 pKa = 3.71ARR616 pKa = 11.84VSFWRR621 pKa = 11.84AFGITPDD628 pKa = 3.86LQQSLEE634 pKa = 4.18DD635 pKa = 4.1EE636 pKa = 4.32YY637 pKa = 11.88SEE639 pKa = 4.2AVVVAKK645 pKa = 9.03PCPMTFPDD653 pKa = 4.07VLAIDD658 pKa = 3.81TSEE661 pKa = 4.33NPLATWLVAARR672 pKa = 11.84TPTLL676 pKa = 3.38
MM1 pKa = 7.12VSAAFTFGCDD11 pKa = 2.81VGTSILLLVAVVGVVLGVRR30 pKa = 11.84WLRR33 pKa = 11.84SSTTGATLVAEE44 pKa = 4.51VDD46 pKa = 4.67LGPDD50 pKa = 3.06WTSVGVTTPYY60 pKa = 10.36QRR62 pKa = 11.84QLAYY66 pKa = 10.24QFKK69 pKa = 11.11AEE71 pKa = 4.13FGEE74 pKa = 4.25LRR76 pKa = 11.84YY77 pKa = 10.71NKK79 pKa = 10.0ANRR82 pKa = 11.84IIAGDD87 pKa = 3.6WCRR90 pKa = 11.84KK91 pKa = 9.38AMADD95 pKa = 3.15NDD97 pKa = 3.51VRR99 pKa = 11.84LTDD102 pKa = 3.73RR103 pKa = 11.84VRR105 pKa = 11.84LLPLAVEE112 pKa = 4.7LCLLPTRR119 pKa = 11.84HH120 pKa = 5.83AVAAAEE126 pKa = 3.89LAATHH131 pKa = 5.78EE132 pKa = 4.41VRR134 pKa = 11.84HH135 pKa = 5.7RR136 pKa = 11.84RR137 pKa = 11.84AAADD141 pKa = 3.81LPKK144 pKa = 9.71XGCPAILPGVTTRR157 pKa = 11.84VDD159 pKa = 3.51RR160 pKa = 11.84VGEE163 pKa = 3.76ACLRR167 pKa = 11.84VRR169 pKa = 11.84EE170 pKa = 4.23CVGVGVRR177 pKa = 11.84SGVDD181 pKa = 2.86RR182 pKa = 11.84SVRR185 pKa = 11.84YY186 pKa = 7.99MAGFGTGVRR195 pKa = 11.84YY196 pKa = 9.18GVHH199 pKa = 6.34CANLKK204 pKa = 9.66NLARR208 pKa = 11.84GIVEE212 pKa = 4.08RR213 pKa = 11.84VFHH216 pKa = 5.21VVRR219 pKa = 11.84DD220 pKa = 3.9GRR222 pKa = 11.84LAQAPQPRR230 pKa = 11.84FGVFSRR236 pKa = 11.84LNSVRR241 pKa = 11.84QRR243 pKa = 11.84LLAVTRR249 pKa = 11.84PTPVVARR256 pKa = 11.84EE257 pKa = 4.52DD258 pKa = 3.89YY259 pKa = 10.25PGLYY263 pKa = 9.31TGRR266 pKa = 11.84KK267 pKa = 7.19RR268 pKa = 11.84GIYY271 pKa = 9.33EE272 pKa = 3.9RR273 pKa = 11.84ALEE276 pKa = 4.21SLTVRR281 pKa = 11.84AINSRR286 pKa = 11.84DD287 pKa = 2.83AWVNTFVKK295 pKa = 10.53AEE297 pKa = 4.02KK298 pKa = 10.82VNLDD302 pKa = 3.82SKK304 pKa = 11.25GDD306 pKa = 3.66PAPRR310 pKa = 11.84VIQPRR315 pKa = 11.84SPRR318 pKa = 11.84YY319 pKa = 7.87NLEE322 pKa = 3.47VGRR325 pKa = 11.84YY326 pKa = 7.88LKK328 pKa = 10.18MFEE331 pKa = 5.17RR332 pKa = 11.84EE333 pKa = 3.93LCHH336 pKa = 6.7GFEE339 pKa = 4.6RR340 pKa = 11.84VWGYY344 pKa = 9.87PVVLKK349 pKa = 10.79GMNAQQVGGWMAAHH363 pKa = 7.16WGAFRR368 pKa = 11.84RR369 pKa = 11.84PVAVGLDD376 pKa = 3.23ASRR379 pKa = 11.84FDD381 pKa = 3.33QHH383 pKa = 7.36VSYY386 pKa = 11.07DD387 pKa = 3.46ALRR390 pKa = 11.84WEE392 pKa = 4.12HH393 pKa = 6.33SVYY396 pKa = 11.01NSTFHH401 pKa = 7.78SPEE404 pKa = 3.71LAKK407 pKa = 10.7LLRR410 pKa = 11.84WQLHH414 pKa = 4.33NRR416 pKa = 11.84GVARR420 pKa = 11.84TEE422 pKa = 4.41GYY424 pKa = 10.43RR425 pKa = 11.84IDD427 pKa = 3.8YY428 pKa = 9.84DD429 pKa = 3.33IRR431 pKa = 11.84GCRR434 pKa = 11.84MSGDD438 pKa = 3.53INTGMGNCLLMSSMVIAYY456 pKa = 9.09CEE458 pKa = 4.22SVGIEE463 pKa = 3.61YY464 pKa = 10.82RR465 pKa = 11.84LANNGDD471 pKa = 3.67DD472 pKa = 3.37CVLFVEE478 pKa = 4.57QQDD481 pKa = 4.05LVKK484 pKa = 10.93LSGIDD489 pKa = 3.07EE490 pKa = 4.19WMLDD494 pKa = 3.49FGFTLTRR501 pKa = 11.84EE502 pKa = 3.91EE503 pKa = 3.77PAYY506 pKa = 10.54VLEE509 pKa = 4.59HH510 pKa = 7.73VEE512 pKa = 4.54FCQARR517 pKa = 11.84PVFTSTGWRR526 pKa = 11.84MVRR529 pKa = 11.84DD530 pKa = 3.53PRR532 pKa = 11.84VAMSKK537 pKa = 10.67DD538 pKa = 3.69CVSLLGWEE546 pKa = 4.41SDD548 pKa = 3.87LDD550 pKa = 3.79LQYY553 pKa = 10.15WCHH556 pKa = 7.41AIGTCGASLTAGVPVWHH573 pKa = 6.47SWYY576 pKa = 10.3HH577 pKa = 5.22RR578 pKa = 11.84LQRR581 pKa = 11.84MGAVAPSGVVDD592 pKa = 3.57MVYY595 pKa = 10.95DD596 pKa = 3.56SGLGYY601 pKa = 9.75MSRR604 pKa = 11.84GVVGGEE610 pKa = 3.8VCAEE614 pKa = 3.71ARR616 pKa = 11.84VSFWRR621 pKa = 11.84AFGITPDD628 pKa = 3.86LQQSLEE634 pKa = 4.18DD635 pKa = 4.1EE636 pKa = 4.32YY637 pKa = 11.88SEE639 pKa = 4.2AVVVAKK645 pKa = 9.03PCPMTFPDD653 pKa = 4.07VLAIDD658 pKa = 3.81TSEE661 pKa = 4.33NPLATWLVAARR672 pKa = 11.84TPTLL676 pKa = 3.38
Molecular weight: 75.15 kDa
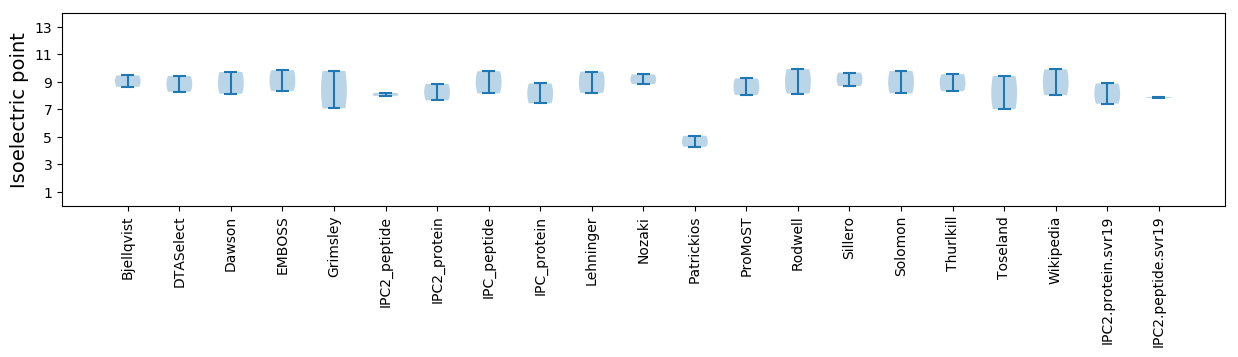
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH37|A0A1L3KH37_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 1 OX=1923662 PE=3 SV=1
MM1 pKa = 7.32ARR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 9.36NPNPVTGGVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84TRR21 pKa = 11.84APNMKK26 pKa = 9.68GGEE29 pKa = 3.93AGTIVEE35 pKa = 4.32YY36 pKa = 10.77SAIGATVTTGGTGLANHH53 pKa = 6.45KK54 pKa = 10.01RR55 pKa = 11.84VFIGGSPYY63 pKa = 11.26DD64 pKa = 3.6LTNTVGPSICSYY76 pKa = 11.28YY77 pKa = 9.34STCKK81 pKa = 10.54FIPGTKK87 pKa = 9.22IRR89 pKa = 11.84WEE91 pKa = 3.99PSVSFTTPGRR101 pKa = 11.84VYY103 pKa = 11.05VGFTDD108 pKa = 4.26NPEE111 pKa = 3.98TMTNIQSAATQADD124 pKa = 3.76WNNLVKK130 pKa = 10.89GLGDD134 pKa = 3.7VISFPVWQEE143 pKa = 3.68TEE145 pKa = 3.9INFPTKK151 pKa = 10.3LRR153 pKa = 11.84RR154 pKa = 11.84KK155 pKa = 9.3MFDD158 pKa = 3.07TNSAVSFVDD167 pKa = 3.97VNALDD172 pKa = 4.23RR173 pKa = 11.84SAQIGMFVAVDD184 pKa = 4.11GAPVTTSVGSFWYY197 pKa = 9.45HH198 pKa = 6.75DD199 pKa = 3.52KK200 pKa = 11.44VLVEE204 pKa = 5.07GIQPTLTT211 pKa = 3.35
MM1 pKa = 7.32ARR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 9.36NPNPVTGGVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84TRR21 pKa = 11.84APNMKK26 pKa = 9.68GGEE29 pKa = 3.93AGTIVEE35 pKa = 4.32YY36 pKa = 10.77SAIGATVTTGGTGLANHH53 pKa = 6.45KK54 pKa = 10.01RR55 pKa = 11.84VFIGGSPYY63 pKa = 11.26DD64 pKa = 3.6LTNTVGPSICSYY76 pKa = 11.28YY77 pKa = 9.34STCKK81 pKa = 10.54FIPGTKK87 pKa = 9.22IRR89 pKa = 11.84WEE91 pKa = 3.99PSVSFTTPGRR101 pKa = 11.84VYY103 pKa = 11.05VGFTDD108 pKa = 4.26NPEE111 pKa = 3.98TMTNIQSAATQADD124 pKa = 3.76WNNLVKK130 pKa = 10.89GLGDD134 pKa = 3.7VISFPVWQEE143 pKa = 3.68TEE145 pKa = 3.9INFPTKK151 pKa = 10.3LRR153 pKa = 11.84RR154 pKa = 11.84KK155 pKa = 9.3MFDD158 pKa = 3.07TNSAVSFVDD167 pKa = 3.97VNALDD172 pKa = 4.23RR173 pKa = 11.84SAQIGMFVAVDD184 pKa = 4.11GAPVTTSVGSFWYY197 pKa = 9.45HH198 pKa = 6.75DD199 pKa = 3.52KK200 pKa = 11.44VLVEE204 pKa = 5.07GIQPTLTT211 pKa = 3.35
Molecular weight: 22.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
887 |
211 |
676 |
443.5 |
49.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.681 ± 0.976 | 2.255 ± 0.623 |
5.073 ± 0.385 | 4.735 ± 0.676 |
3.608 ± 0.54 | 8.794 ± 0.553 |
1.917 ± 0.462 | 3.157 ± 0.981 |
2.706 ± 0.744 | 7.892 ± 1.955 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.48 ± 0.053 | 3.382 ± 1.099 |
4.848 ± 0.626 | 2.593 ± 0.106 |
8.681 ± 1.202 | 5.75 ± 0.648 |
6.539 ± 2.306 | 11.161 ± 0.576 |
2.368 ± 0.225 | 3.269 ± 0.203 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
