
Wuchang romanomermis nematode virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Lispiviridae; Arlivirus; Wuchang arlivirus
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
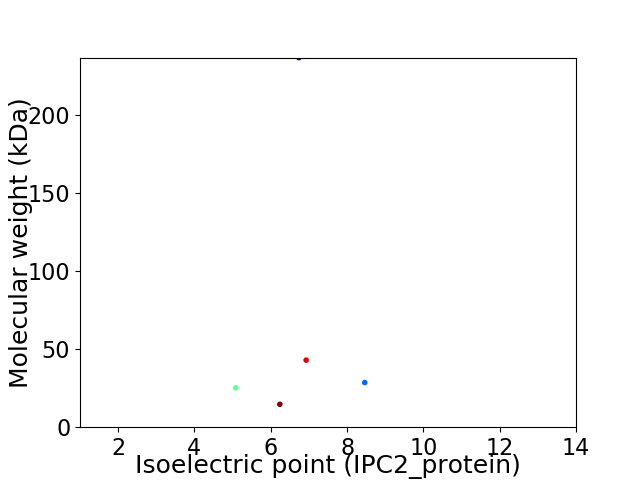
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KN23|A0A1L3KN23_9MONO RNA-directed RNA polymerase L OS=Wuchang romanomermis nematode virus 2 OX=2773460 PE=3 SV=1
MM1 pKa = 8.36SEE3 pKa = 4.05EE4 pKa = 4.54NLNEE8 pKa = 4.06MPNGAEE14 pKa = 4.07AAMKK18 pKa = 10.35ANNLIRR24 pKa = 11.84MTRR27 pKa = 11.84NQSMAADD34 pKa = 4.11LLEE37 pKa = 3.79QQYY40 pKa = 10.59RR41 pKa = 11.84GEE43 pKa = 4.58LDD45 pKa = 4.76PILEE49 pKa = 4.44TPTEE53 pKa = 4.22YY54 pKa = 10.56EE55 pKa = 4.17DD56 pKa = 4.1KK57 pKa = 11.33VDD59 pKa = 3.79VSLQALVAINGKK71 pKa = 9.15LADD74 pKa = 4.08VLNGLQMVTTSTQLIMEE91 pKa = 5.69NIPDD95 pKa = 3.86VKK97 pKa = 10.66DD98 pKa = 3.59GNQMKK103 pKa = 8.75EE104 pKa = 4.17TVLTDD109 pKa = 3.91LKK111 pKa = 11.41AHH113 pKa = 6.58MSQEE117 pKa = 3.69IVALYY122 pKa = 10.68ARR124 pKa = 11.84LSKK127 pKa = 10.6EE128 pKa = 3.63IADD131 pKa = 3.76MKK133 pKa = 10.47QAFIGEE139 pKa = 4.18LQKK142 pKa = 10.48IRR144 pKa = 11.84TEE146 pKa = 3.72IQALKK151 pKa = 8.92GQKK154 pKa = 10.43SMASLSLQPVSQPMVATTSSAVMPTEE180 pKa = 3.52KK181 pKa = 10.37RR182 pKa = 11.84KK183 pKa = 9.92IDD185 pKa = 3.28MRR187 pKa = 11.84EE188 pKa = 3.6YY189 pKa = 11.25SEE191 pKa = 4.17ILSAFRR197 pKa = 11.84NYY199 pKa = 10.15EE200 pKa = 3.61GSDD203 pKa = 3.37ANKK206 pKa = 10.04QAIQAAILRR215 pKa = 11.84RR216 pKa = 11.84DD217 pKa = 3.58LVAARR222 pKa = 11.84AIMNGG227 pKa = 3.18
MM1 pKa = 8.36SEE3 pKa = 4.05EE4 pKa = 4.54NLNEE8 pKa = 4.06MPNGAEE14 pKa = 4.07AAMKK18 pKa = 10.35ANNLIRR24 pKa = 11.84MTRR27 pKa = 11.84NQSMAADD34 pKa = 4.11LLEE37 pKa = 3.79QQYY40 pKa = 10.59RR41 pKa = 11.84GEE43 pKa = 4.58LDD45 pKa = 4.76PILEE49 pKa = 4.44TPTEE53 pKa = 4.22YY54 pKa = 10.56EE55 pKa = 4.17DD56 pKa = 4.1KK57 pKa = 11.33VDD59 pKa = 3.79VSLQALVAINGKK71 pKa = 9.15LADD74 pKa = 4.08VLNGLQMVTTSTQLIMEE91 pKa = 5.69NIPDD95 pKa = 3.86VKK97 pKa = 10.66DD98 pKa = 3.59GNQMKK103 pKa = 8.75EE104 pKa = 4.17TVLTDD109 pKa = 3.91LKK111 pKa = 11.41AHH113 pKa = 6.58MSQEE117 pKa = 3.69IVALYY122 pKa = 10.68ARR124 pKa = 11.84LSKK127 pKa = 10.6EE128 pKa = 3.63IADD131 pKa = 3.76MKK133 pKa = 10.47QAFIGEE139 pKa = 4.18LQKK142 pKa = 10.48IRR144 pKa = 11.84TEE146 pKa = 3.72IQALKK151 pKa = 8.92GQKK154 pKa = 10.43SMASLSLQPVSQPMVATTSSAVMPTEE180 pKa = 3.52KK181 pKa = 10.37RR182 pKa = 11.84KK183 pKa = 9.92IDD185 pKa = 3.28MRR187 pKa = 11.84EE188 pKa = 3.6YY189 pKa = 11.25SEE191 pKa = 4.17ILSAFRR197 pKa = 11.84NYY199 pKa = 10.15EE200 pKa = 3.61GSDD203 pKa = 3.37ANKK206 pKa = 10.04QAIQAAILRR215 pKa = 11.84RR216 pKa = 11.84DD217 pKa = 3.58LVAARR222 pKa = 11.84AIMNGG227 pKa = 3.18
Molecular weight: 25.16 kDa
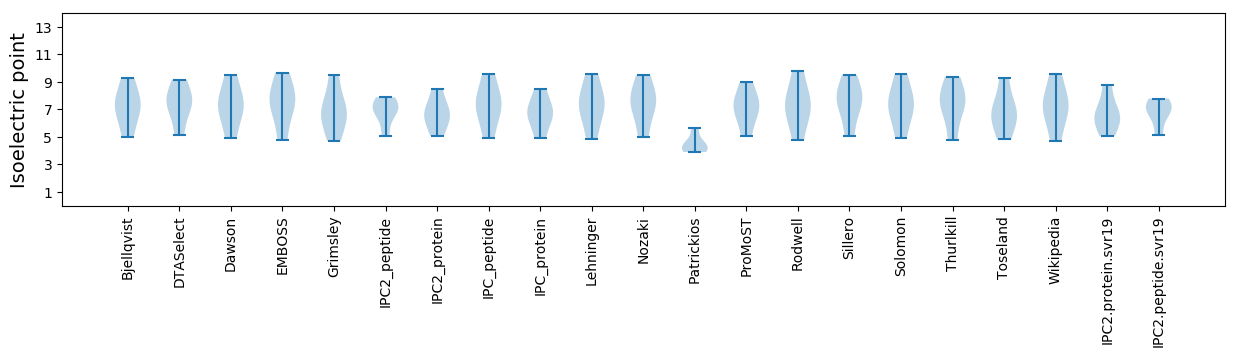
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KN03|A0A1L3KN03_9MONO Uncharacterized protein OS=Wuchang romanomermis nematode virus 2 OX=2773460 PE=4 SV=1
MM1 pKa = 7.94DD2 pKa = 5.38RR3 pKa = 11.84PTFTRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84SSSVGPTIEE19 pKa = 3.95SRR21 pKa = 11.84PTVPCKK27 pKa = 10.16YY28 pKa = 9.63WEE30 pKa = 4.78DD31 pKa = 3.16SHH33 pKa = 8.68ICMNGASCRR42 pKa = 11.84FAHH45 pKa = 6.79NDD47 pKa = 3.45YY48 pKa = 11.26DD49 pKa = 4.16PTPLDD54 pKa = 3.21IVRR57 pKa = 11.84HH58 pKa = 5.55LDD60 pKa = 3.51FMVGIMNKK68 pKa = 6.92ITEE71 pKa = 4.69RR72 pKa = 11.84ISRR75 pKa = 11.84LEE77 pKa = 3.82MKK79 pKa = 10.65VSGRR83 pKa = 11.84SNFQQQGTLQQRR95 pKa = 11.84ASRR98 pKa = 11.84MLAHH102 pKa = 7.05SNRR105 pKa = 11.84FDD107 pKa = 3.28RR108 pKa = 11.84RR109 pKa = 11.84SRR111 pKa = 11.84YY112 pKa = 8.74HH113 pKa = 7.04ASPSSAVSRR122 pKa = 11.84MSQRR126 pKa = 11.84DD127 pKa = 3.79EE128 pKa = 4.63LDD130 pKa = 2.94SSQPVQTSFRR140 pKa = 11.84TEE142 pKa = 3.52PHH144 pKa = 6.79HH145 pKa = 7.25NPFSPDD151 pKa = 2.92LMGQFTPQIPSFSYY165 pKa = 8.56QQAWEE170 pKa = 4.24MPPKK174 pKa = 10.52PFVNKK179 pKa = 8.43NTQIAKK185 pKa = 10.21KK186 pKa = 9.88IAEE189 pKa = 4.16EE190 pKa = 3.83KK191 pKa = 9.25QANIKK196 pKa = 10.74AMLEE200 pKa = 4.03LEE202 pKa = 4.87KK203 pKa = 10.68RR204 pKa = 11.84LQNNLDD210 pKa = 3.29QATVKK215 pKa = 10.68NVLKK219 pKa = 10.77NAGPSTDD226 pKa = 3.44SQIDD230 pKa = 3.46SDD232 pKa = 4.0KK233 pKa = 11.07KK234 pKa = 10.18QEE236 pKa = 4.03KK237 pKa = 10.38EE238 pKa = 3.86DD239 pKa = 3.76MKK241 pKa = 11.26QEE243 pKa = 4.25GQSEE247 pKa = 4.33FF248 pKa = 3.67
MM1 pKa = 7.94DD2 pKa = 5.38RR3 pKa = 11.84PTFTRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84SSSVGPTIEE19 pKa = 3.95SRR21 pKa = 11.84PTVPCKK27 pKa = 10.16YY28 pKa = 9.63WEE30 pKa = 4.78DD31 pKa = 3.16SHH33 pKa = 8.68ICMNGASCRR42 pKa = 11.84FAHH45 pKa = 6.79NDD47 pKa = 3.45YY48 pKa = 11.26DD49 pKa = 4.16PTPLDD54 pKa = 3.21IVRR57 pKa = 11.84HH58 pKa = 5.55LDD60 pKa = 3.51FMVGIMNKK68 pKa = 6.92ITEE71 pKa = 4.69RR72 pKa = 11.84ISRR75 pKa = 11.84LEE77 pKa = 3.82MKK79 pKa = 10.65VSGRR83 pKa = 11.84SNFQQQGTLQQRR95 pKa = 11.84ASRR98 pKa = 11.84MLAHH102 pKa = 7.05SNRR105 pKa = 11.84FDD107 pKa = 3.28RR108 pKa = 11.84RR109 pKa = 11.84SRR111 pKa = 11.84YY112 pKa = 8.74HH113 pKa = 7.04ASPSSAVSRR122 pKa = 11.84MSQRR126 pKa = 11.84DD127 pKa = 3.79EE128 pKa = 4.63LDD130 pKa = 2.94SSQPVQTSFRR140 pKa = 11.84TEE142 pKa = 3.52PHH144 pKa = 6.79HH145 pKa = 7.25NPFSPDD151 pKa = 2.92LMGQFTPQIPSFSYY165 pKa = 8.56QQAWEE170 pKa = 4.24MPPKK174 pKa = 10.52PFVNKK179 pKa = 8.43NTQIAKK185 pKa = 10.21KK186 pKa = 9.88IAEE189 pKa = 4.16EE190 pKa = 3.83KK191 pKa = 9.25QANIKK196 pKa = 10.74AMLEE200 pKa = 4.03LEE202 pKa = 4.87KK203 pKa = 10.68RR204 pKa = 11.84LQNNLDD210 pKa = 3.29QATVKK215 pKa = 10.68NVLKK219 pKa = 10.77NAGPSTDD226 pKa = 3.44SQIDD230 pKa = 3.46SDD232 pKa = 4.0KK233 pKa = 11.07KK234 pKa = 10.18QEE236 pKa = 4.03KK237 pKa = 10.38EE238 pKa = 3.86DD239 pKa = 3.76MKK241 pKa = 11.26QEE243 pKa = 4.25GQSEE247 pKa = 4.33FF248 pKa = 3.67
Molecular weight: 28.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3045 |
129 |
2058 |
609.0 |
69.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.568 ± 0.858 | 1.609 ± 0.325 |
5.156 ± 0.257 | 5.846 ± 0.428 |
4.302 ± 0.589 | 4.039 ± 0.233 |
2.266 ± 0.354 | 7.98 ± 0.889 |
5.944 ± 0.143 | 10.608 ± 1.179 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.791 ± 0.797 | 4.532 ± 0.475 |
4.532 ± 0.542 | 4.368 ± 0.98 |
5.09 ± 0.598 | 7.258 ± 0.717 |
6.305 ± 0.321 | 5.484 ± 0.286 |
1.478 ± 0.423 | 3.842 ± 0.642 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
