
Tenacibaculum maritimum NCIMB 2154
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum; Tenacibaculum maritimum
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
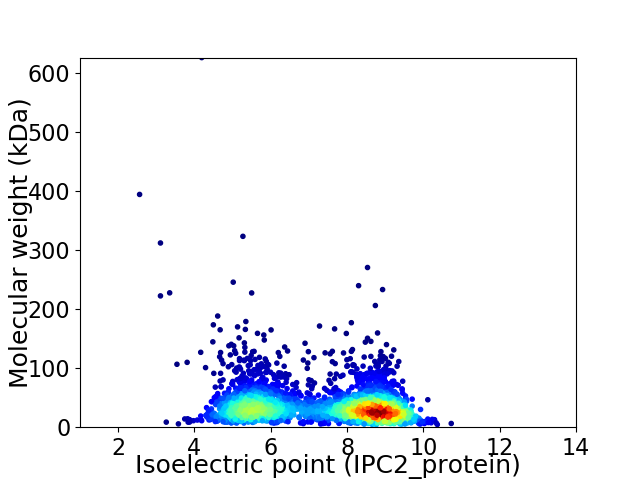
Virtual 2D-PAGE plot for 2844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H1E8D9|A0A2H1E8D9_9FLAO 4-hydroxy-tetrahydrodipicolinate reductase OS=Tenacibaculum maritimum NCIMB 2154 OX=1349785 GN=dapB PE=3 SV=1
MM1 pKa = 7.42IKK3 pKa = 10.13NYY5 pKa = 10.75SMSILKK11 pKa = 10.36LNSEE15 pKa = 4.55IKK17 pKa = 9.44STATFLLLLLLAGNFMYY34 pKa = 9.66GQCTDD39 pKa = 3.33GEE41 pKa = 5.13LNFTWSGQAGGGNQFVWNAGQRR63 pKa = 11.84TGSDD67 pKa = 3.12TFTSCASNVTVTVTVDD83 pKa = 3.29DD84 pKa = 5.05DD85 pKa = 4.59NNGSSEE91 pKa = 4.18VYY93 pKa = 10.64YY94 pKa = 10.85DD95 pKa = 4.25GSQSGGNSTSGGGAFANQGLTIWLDD120 pKa = 3.4NNEE123 pKa = 4.38NLFDD127 pKa = 4.76ADD129 pKa = 3.45AMVVGEE135 pKa = 4.25RR136 pKa = 11.84ATIVFDD142 pKa = 3.81FSVPVEE148 pKa = 4.14LKK150 pKa = 10.83SFLTGDD156 pKa = 3.62IDD158 pKa = 3.72WNNPGNISSTWRR170 pKa = 11.84DD171 pKa = 3.62FVRR174 pKa = 11.84VSASNGGTNVPVQGIVQAPGTVTMVGQLAASNLGAIGSTGGLDD217 pKa = 3.37PSDD220 pKa = 3.87PRR222 pKa = 11.84GHH224 pKa = 6.57VIWNSQGNLITSLRR238 pKa = 11.84VTYY241 pKa = 9.98EE242 pKa = 3.76VGVSGTGQQSLLIGSFLFDD261 pKa = 4.4ADD263 pKa = 3.76ADD265 pKa = 4.07NDD267 pKa = 4.15GVLNDD272 pKa = 3.52VDD274 pKa = 3.66QCEE277 pKa = 4.33GFDD280 pKa = 5.23DD281 pKa = 5.62NMDD284 pKa = 3.95ADD286 pKa = 4.49GDD288 pKa = 4.47GIADD292 pKa = 4.16GCDD295 pKa = 3.08LDD297 pKa = 5.36ADD299 pKa = 3.92NDD301 pKa = 4.71GILNVDD307 pKa = 4.4EE308 pKa = 4.73YY309 pKa = 11.18NNCITPTIEE318 pKa = 3.78LKK320 pKa = 10.66KK321 pKa = 10.99YY322 pKa = 10.17FLDD325 pKa = 4.7DD326 pKa = 4.49FGQGTVRR333 pKa = 11.84TSTPYY338 pKa = 10.11ISNSYY343 pKa = 9.63FFNSIADD350 pKa = 3.85VNDD353 pKa = 3.66GQYY356 pKa = 11.1CVIDD360 pKa = 3.95TPNPGAGNHH369 pKa = 5.45VSWGTYY375 pKa = 9.27GDD377 pKa = 3.71HH378 pKa = 7.47TIGDD382 pKa = 4.31TNGKK386 pKa = 8.25MLVINSSSSTFAHH399 pKa = 6.2YY400 pKa = 10.36YY401 pKa = 8.99KK402 pKa = 10.6RR403 pKa = 11.84SIEE406 pKa = 4.07NIIPNTIVKK415 pKa = 10.45LSFWARR421 pKa = 11.84NICAGCTSQPDD432 pKa = 3.33ITYY435 pKa = 10.5SILDD439 pKa = 3.35SSDD442 pKa = 3.43NVLATANTGGLSTSDD457 pKa = 2.72WVNYY461 pKa = 9.44EE462 pKa = 3.68LTVNIGSDD470 pKa = 3.18TDD472 pKa = 3.89LQISLTNNNVPSGDD486 pKa = 3.96GNDD489 pKa = 3.96LAIDD493 pKa = 4.96DD494 pKa = 4.77ISFSSIVCDD503 pKa = 3.78SDD505 pKa = 4.54SDD507 pKa = 4.3TIPNYY512 pKa = 10.88LDD514 pKa = 3.26VDD516 pKa = 4.33SDD518 pKa = 4.29NDD520 pKa = 3.92GCPDD524 pKa = 3.28AMEE527 pKa = 5.03GGNTGLNLTDD537 pKa = 5.5LNILNQLVGGVDD549 pKa = 3.89TNSSSASYY557 pKa = 10.01GVPIIAGGGQVDD569 pKa = 3.99VSSTNASVQSAEE581 pKa = 4.81CNPCDD586 pKa = 3.98ATSTLYY592 pKa = 10.75SDD594 pKa = 4.74RR595 pKa = 11.84DD596 pKa = 3.41GDD598 pKa = 4.19NVGDD602 pKa = 4.42ACDD605 pKa = 4.66SDD607 pKa = 4.55DD608 pKa = 5.62DD609 pKa = 4.53NDD611 pKa = 5.3GILDD615 pKa = 3.96EE616 pKa = 5.84NEE618 pKa = 4.17GCPSNFTPGEE628 pKa = 3.95VRR630 pKa = 11.84LIPPVGAWEE639 pKa = 4.02VFLYY643 pKa = 10.54DD644 pKa = 4.91GRR646 pKa = 11.84FQVLNALNLVDD657 pKa = 4.06VPSRR661 pKa = 11.84GADD664 pKa = 3.45GTPGGLVVLSAHH676 pKa = 6.57GFYY679 pKa = 10.13TGNVGTYY686 pKa = 7.61TFNDD690 pKa = 3.32VSIGANQDD698 pKa = 3.19PTPSVVASGVDD709 pKa = 3.48MKK711 pKa = 11.35VLTPFTVSNSGDD723 pKa = 2.72WSIVYY728 pKa = 10.03KK729 pKa = 9.91RR730 pKa = 11.84TIEE733 pKa = 4.02NNGTITIGAPGSYY746 pKa = 10.09VDD748 pKa = 4.87DD749 pKa = 4.65WIEE752 pKa = 3.77LFINGVLVDD761 pKa = 4.09SVNSFTASLPVADD774 pKa = 5.1VISEE778 pKa = 4.23NISAGDD784 pKa = 3.57VVEE787 pKa = 4.43VRR789 pKa = 11.84LTNGLGLGGFNLSIQTLNEE808 pKa = 3.39VTLAVVCDD816 pKa = 3.87TDD818 pKa = 3.38TDD820 pKa = 3.75NDD822 pKa = 3.9GVLNHH827 pKa = 7.35LDD829 pKa = 3.81TDD831 pKa = 4.28SDD833 pKa = 5.03DD834 pKa = 5.28DD835 pKa = 4.21GCPDD839 pKa = 4.48AIEE842 pKa = 4.39GAGNIIGGLSTLTGGSPTAGSSSQNLGTNVDD873 pKa = 3.48AEE875 pKa = 4.59GNPKK879 pKa = 9.6INAGDD884 pKa = 3.59TTGFEE889 pKa = 4.64QNNTAAVIDD898 pKa = 4.04NTDD901 pKa = 3.38STGCTADD908 pKa = 3.56LSLTKK913 pKa = 10.1KK914 pKa = 10.05VNKK917 pKa = 9.9ALPKK921 pKa = 10.07VGDD924 pKa = 4.52DD925 pKa = 2.76IVYY928 pKa = 10.27ILTVKK933 pKa = 10.97NEE935 pKa = 4.07GPLNASGVKK944 pKa = 8.82VTDD947 pKa = 3.55VLPVGLTYY955 pKa = 10.43KK956 pKa = 10.69SSVIDD961 pKa = 4.21LLGTYY966 pKa = 7.89TNNVWDD972 pKa = 3.92IGNLNVGEE980 pKa = 4.38SVNLRR985 pKa = 11.84ITATVTQQGTIINTAEE1001 pKa = 3.95ITQSNQVDD1009 pKa = 3.47IDD1011 pKa = 4.01STPMSGNN1018 pKa = 3.24
MM1 pKa = 7.42IKK3 pKa = 10.13NYY5 pKa = 10.75SMSILKK11 pKa = 10.36LNSEE15 pKa = 4.55IKK17 pKa = 9.44STATFLLLLLLAGNFMYY34 pKa = 9.66GQCTDD39 pKa = 3.33GEE41 pKa = 5.13LNFTWSGQAGGGNQFVWNAGQRR63 pKa = 11.84TGSDD67 pKa = 3.12TFTSCASNVTVTVTVDD83 pKa = 3.29DD84 pKa = 5.05DD85 pKa = 4.59NNGSSEE91 pKa = 4.18VYY93 pKa = 10.64YY94 pKa = 10.85DD95 pKa = 4.25GSQSGGNSTSGGGAFANQGLTIWLDD120 pKa = 3.4NNEE123 pKa = 4.38NLFDD127 pKa = 4.76ADD129 pKa = 3.45AMVVGEE135 pKa = 4.25RR136 pKa = 11.84ATIVFDD142 pKa = 3.81FSVPVEE148 pKa = 4.14LKK150 pKa = 10.83SFLTGDD156 pKa = 3.62IDD158 pKa = 3.72WNNPGNISSTWRR170 pKa = 11.84DD171 pKa = 3.62FVRR174 pKa = 11.84VSASNGGTNVPVQGIVQAPGTVTMVGQLAASNLGAIGSTGGLDD217 pKa = 3.37PSDD220 pKa = 3.87PRR222 pKa = 11.84GHH224 pKa = 6.57VIWNSQGNLITSLRR238 pKa = 11.84VTYY241 pKa = 9.98EE242 pKa = 3.76VGVSGTGQQSLLIGSFLFDD261 pKa = 4.4ADD263 pKa = 3.76ADD265 pKa = 4.07NDD267 pKa = 4.15GVLNDD272 pKa = 3.52VDD274 pKa = 3.66QCEE277 pKa = 4.33GFDD280 pKa = 5.23DD281 pKa = 5.62NMDD284 pKa = 3.95ADD286 pKa = 4.49GDD288 pKa = 4.47GIADD292 pKa = 4.16GCDD295 pKa = 3.08LDD297 pKa = 5.36ADD299 pKa = 3.92NDD301 pKa = 4.71GILNVDD307 pKa = 4.4EE308 pKa = 4.73YY309 pKa = 11.18NNCITPTIEE318 pKa = 3.78LKK320 pKa = 10.66KK321 pKa = 10.99YY322 pKa = 10.17FLDD325 pKa = 4.7DD326 pKa = 4.49FGQGTVRR333 pKa = 11.84TSTPYY338 pKa = 10.11ISNSYY343 pKa = 9.63FFNSIADD350 pKa = 3.85VNDD353 pKa = 3.66GQYY356 pKa = 11.1CVIDD360 pKa = 3.95TPNPGAGNHH369 pKa = 5.45VSWGTYY375 pKa = 9.27GDD377 pKa = 3.71HH378 pKa = 7.47TIGDD382 pKa = 4.31TNGKK386 pKa = 8.25MLVINSSSSTFAHH399 pKa = 6.2YY400 pKa = 10.36YY401 pKa = 8.99KK402 pKa = 10.6RR403 pKa = 11.84SIEE406 pKa = 4.07NIIPNTIVKK415 pKa = 10.45LSFWARR421 pKa = 11.84NICAGCTSQPDD432 pKa = 3.33ITYY435 pKa = 10.5SILDD439 pKa = 3.35SSDD442 pKa = 3.43NVLATANTGGLSTSDD457 pKa = 2.72WVNYY461 pKa = 9.44EE462 pKa = 3.68LTVNIGSDD470 pKa = 3.18TDD472 pKa = 3.89LQISLTNNNVPSGDD486 pKa = 3.96GNDD489 pKa = 3.96LAIDD493 pKa = 4.96DD494 pKa = 4.77ISFSSIVCDD503 pKa = 3.78SDD505 pKa = 4.54SDD507 pKa = 4.3TIPNYY512 pKa = 10.88LDD514 pKa = 3.26VDD516 pKa = 4.33SDD518 pKa = 4.29NDD520 pKa = 3.92GCPDD524 pKa = 3.28AMEE527 pKa = 5.03GGNTGLNLTDD537 pKa = 5.5LNILNQLVGGVDD549 pKa = 3.89TNSSSASYY557 pKa = 10.01GVPIIAGGGQVDD569 pKa = 3.99VSSTNASVQSAEE581 pKa = 4.81CNPCDD586 pKa = 3.98ATSTLYY592 pKa = 10.75SDD594 pKa = 4.74RR595 pKa = 11.84DD596 pKa = 3.41GDD598 pKa = 4.19NVGDD602 pKa = 4.42ACDD605 pKa = 4.66SDD607 pKa = 4.55DD608 pKa = 5.62DD609 pKa = 4.53NDD611 pKa = 5.3GILDD615 pKa = 3.96EE616 pKa = 5.84NEE618 pKa = 4.17GCPSNFTPGEE628 pKa = 3.95VRR630 pKa = 11.84LIPPVGAWEE639 pKa = 4.02VFLYY643 pKa = 10.54DD644 pKa = 4.91GRR646 pKa = 11.84FQVLNALNLVDD657 pKa = 4.06VPSRR661 pKa = 11.84GADD664 pKa = 3.45GTPGGLVVLSAHH676 pKa = 6.57GFYY679 pKa = 10.13TGNVGTYY686 pKa = 7.61TFNDD690 pKa = 3.32VSIGANQDD698 pKa = 3.19PTPSVVASGVDD709 pKa = 3.48MKK711 pKa = 11.35VLTPFTVSNSGDD723 pKa = 2.72WSIVYY728 pKa = 10.03KK729 pKa = 9.91RR730 pKa = 11.84TIEE733 pKa = 4.02NNGTITIGAPGSYY746 pKa = 10.09VDD748 pKa = 4.87DD749 pKa = 4.65WIEE752 pKa = 3.77LFINGVLVDD761 pKa = 4.09SVNSFTASLPVADD774 pKa = 5.1VISEE778 pKa = 4.23NISAGDD784 pKa = 3.57VVEE787 pKa = 4.43VRR789 pKa = 11.84LTNGLGLGGFNLSIQTLNEE808 pKa = 3.39VTLAVVCDD816 pKa = 3.87TDD818 pKa = 3.38TDD820 pKa = 3.75NDD822 pKa = 3.9GVLNHH827 pKa = 7.35LDD829 pKa = 3.81TDD831 pKa = 4.28SDD833 pKa = 5.03DD834 pKa = 5.28DD835 pKa = 4.21GCPDD839 pKa = 4.48AIEE842 pKa = 4.39GAGNIIGGLSTLTGGSPTAGSSSQNLGTNVDD873 pKa = 3.48AEE875 pKa = 4.59GNPKK879 pKa = 9.6INAGDD884 pKa = 3.59TTGFEE889 pKa = 4.64QNNTAAVIDD898 pKa = 4.04NTDD901 pKa = 3.38STGCTADD908 pKa = 3.56LSLTKK913 pKa = 10.1KK914 pKa = 10.05VNKK917 pKa = 9.9ALPKK921 pKa = 10.07VGDD924 pKa = 4.52DD925 pKa = 2.76IVYY928 pKa = 10.27ILTVKK933 pKa = 10.97NEE935 pKa = 4.07GPLNASGVKK944 pKa = 8.82VTDD947 pKa = 3.55VLPVGLTYY955 pKa = 10.43KK956 pKa = 10.69SSVIDD961 pKa = 4.21LLGTYY966 pKa = 7.89TNNVWDD972 pKa = 3.92IGNLNVGEE980 pKa = 4.38SVNLRR985 pKa = 11.84ITATVTQQGTIINTAEE1001 pKa = 3.95ITQSNQVDD1009 pKa = 3.47IDD1011 pKa = 4.01STPMSGNN1018 pKa = 3.24
Molecular weight: 106.56 kDa
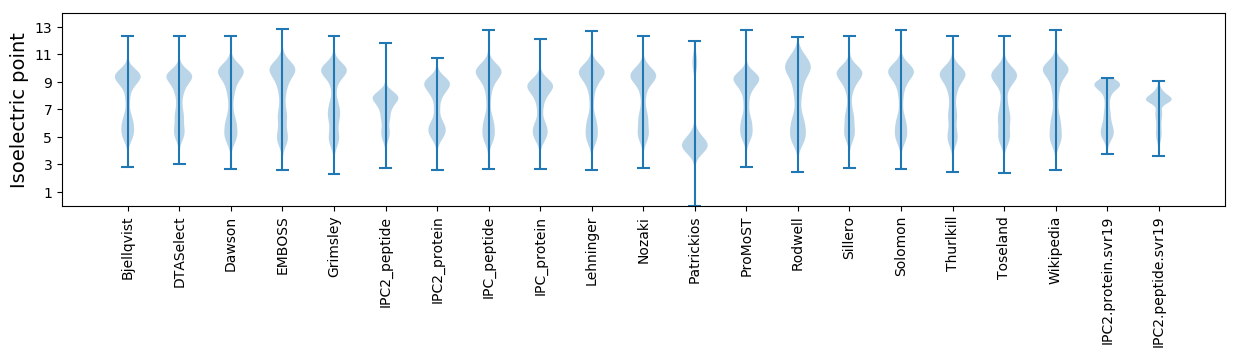
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H1EBK8|A0A2H1EBK8_9FLAO Trk system potassium uptake protein TrkA OS=Tenacibaculum maritimum NCIMB 2154 OX=1349785 GN=trkA PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 3.92TITTDD27 pKa = 3.73KK28 pKa = 10.69PEE30 pKa = 4.27KK31 pKa = 10.46SLLVPKK37 pKa = 10.16KK38 pKa = 10.51RR39 pKa = 11.84SGGRR43 pKa = 11.84NNQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.58KK59 pKa = 9.49QKK61 pKa = 10.8YY62 pKa = 9.58RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.83RR69 pKa = 11.84DD70 pKa = 3.38KK71 pKa = 10.89QGIPATVKK79 pKa = 10.01TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALVSYY96 pKa = 11.47ADD98 pKa = 3.44GEE100 pKa = 4.21KK101 pKa = 10.25RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.19VGQTIISGKK120 pKa = 9.71NVAPEE125 pKa = 4.07IGNAMYY131 pKa = 10.77LSDD134 pKa = 4.56IPLGTTISCIEE145 pKa = 4.05LRR147 pKa = 11.84PEE149 pKa = 3.6QGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.4ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 3.92TITTDD27 pKa = 3.73KK28 pKa = 10.69PEE30 pKa = 4.27KK31 pKa = 10.46SLLVPKK37 pKa = 10.16KK38 pKa = 10.51RR39 pKa = 11.84SGGRR43 pKa = 11.84NNQGRR48 pKa = 11.84MTTRR52 pKa = 11.84NIGGGHH58 pKa = 4.58KK59 pKa = 9.49QKK61 pKa = 10.8YY62 pKa = 9.58RR63 pKa = 11.84IIDD66 pKa = 3.85FKK68 pKa = 10.83RR69 pKa = 11.84DD70 pKa = 3.38KK71 pKa = 10.89QGIPATVKK79 pKa = 10.01TIEE82 pKa = 4.0YY83 pKa = 9.86DD84 pKa = 3.47PNRR87 pKa = 11.84TAFIALVSYY96 pKa = 11.47ADD98 pKa = 3.44GEE100 pKa = 4.21KK101 pKa = 10.25RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.19VGQTIISGKK120 pKa = 9.71NVAPEE125 pKa = 4.07IGNAMYY131 pKa = 10.77LSDD134 pKa = 4.56IPLGTTISCIEE145 pKa = 4.05LRR147 pKa = 11.84PEE149 pKa = 3.6QGAVMARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.52YY171 pKa = 9.12ATVKK175 pKa = 10.42LPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILLTCMATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.49AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TNAVRR224 pKa = 11.84MNPVDD229 pKa = 3.4HH230 pKa = 7.07PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.4ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 30.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
983416 |
38 |
5927 |
345.8 |
39.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.107 ± 0.052 | 0.757 ± 0.017 |
5.063 ± 0.049 | 6.559 ± 0.048 |
5.191 ± 0.038 | 6.235 ± 0.05 |
1.847 ± 0.023 | 8.378 ± 0.04 |
8.989 ± 0.073 | 9.119 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.0 ± 0.024 | 6.39 ± 0.048 |
3.25 ± 0.023 | 3.182 ± 0.024 |
3.375 ± 0.034 | 6.394 ± 0.039 |
5.978 ± 0.073 | 5.938 ± 0.047 |
0.997 ± 0.015 | 4.252 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
