
Beet western yellows virus associated RNA
Taxonomy: Viruses; Riboviria; Virus-associated RNAs
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
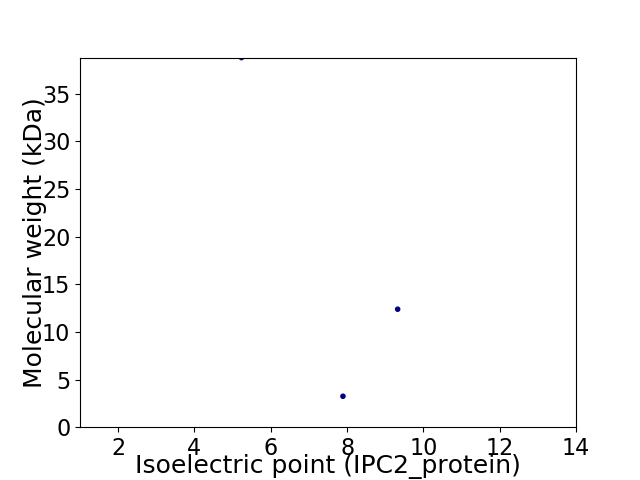
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0P708|A0A0A0P708_9VIRU RNA-directed RNA polymerase OS=Beet western yellows virus associated RNA OX=1425367 PE=4 SV=1
MM1 pKa = 7.67FKK3 pKa = 10.66RR4 pKa = 11.84FTVSDD9 pKa = 3.72TPVCFKK15 pKa = 10.74GLNYY19 pKa = 10.02LRR21 pKa = 11.84RR22 pKa = 11.84GNFMKK27 pKa = 10.53EE28 pKa = 3.27KK29 pKa = 9.53WEE31 pKa = 4.29SFNDD35 pKa = 3.73PVAICLDD42 pKa = 3.45ASRR45 pKa = 11.84FDD47 pKa = 3.57LHH49 pKa = 8.19VSEE52 pKa = 6.09DD53 pKa = 4.03ALQHH57 pKa = 4.62THH59 pKa = 6.62LLYY62 pKa = 8.43MTAIGNDD69 pKa = 3.42PQLAYY74 pKa = 9.79ILEE77 pKa = 4.35NRR79 pKa = 11.84LVTDD83 pKa = 4.44GVGWSSDD90 pKa = 3.07GAFRR94 pKa = 11.84YY95 pKa = 8.82HH96 pKa = 7.1KK97 pKa = 10.64KK98 pKa = 10.54GGRR101 pKa = 11.84CSGDD105 pKa = 2.94NDD107 pKa = 3.61TSLGNVIIMLSITYY121 pKa = 8.74AFCEE125 pKa = 4.27EE126 pKa = 4.06SDD128 pKa = 3.54IPHH131 pKa = 6.82IEE133 pKa = 4.0VTNDD137 pKa = 2.82GDD139 pKa = 3.79DD140 pKa = 3.42QVIMVEE146 pKa = 4.21RR147 pKa = 11.84EE148 pKa = 3.84HH149 pKa = 6.13MHH151 pKa = 5.68TVLQIEE157 pKa = 4.22DD158 pKa = 3.8TFRR161 pKa = 11.84KK162 pKa = 9.75FGFLLKK168 pKa = 10.74VEE170 pKa = 4.4DD171 pKa = 3.93PVYY174 pKa = 9.98EE175 pKa = 3.96LEE177 pKa = 6.09RR178 pKa = 11.84IDD180 pKa = 4.18FCQTRR185 pKa = 11.84PVQLTPDD192 pKa = 3.24VCTMVRR198 pKa = 11.84HH199 pKa = 6.23PRR201 pKa = 11.84LAMTKK206 pKa = 10.27DD207 pKa = 3.25LTTFIPIEE215 pKa = 4.15RR216 pKa = 11.84GKK218 pKa = 10.76LKK220 pKa = 10.85YY221 pKa = 10.34HH222 pKa = 6.45MLTAIGKK229 pKa = 9.25CGSACYY235 pKa = 10.46DD236 pKa = 4.55DD237 pKa = 5.5IPVLRR242 pKa = 11.84AMYY245 pKa = 10.26RR246 pKa = 11.84KK247 pKa = 9.06MEE249 pKa = 4.84IIGTRR254 pKa = 11.84NKK256 pKa = 10.54GNEE259 pKa = 3.97DD260 pKa = 3.38SWWNSTHH267 pKa = 7.24ADD269 pKa = 3.41PAFRR273 pKa = 11.84TLSSKK278 pKa = 10.81AATNVGLTTQCRR290 pKa = 11.84ISFWKK295 pKa = 10.69AFGILPDD302 pKa = 3.57QQEE305 pKa = 4.02ALEE308 pKa = 4.25NEE310 pKa = 3.79WRR312 pKa = 11.84QWEE315 pKa = 4.06PSLDD319 pKa = 3.11RR320 pKa = 11.84VEE322 pKa = 5.0FVHH325 pKa = 6.78SFLGDD330 pKa = 3.57YY331 pKa = 7.89PVCEE335 pKa = 4.23GDD337 pKa = 3.51VV338 pKa = 3.25
MM1 pKa = 7.67FKK3 pKa = 10.66RR4 pKa = 11.84FTVSDD9 pKa = 3.72TPVCFKK15 pKa = 10.74GLNYY19 pKa = 10.02LRR21 pKa = 11.84RR22 pKa = 11.84GNFMKK27 pKa = 10.53EE28 pKa = 3.27KK29 pKa = 9.53WEE31 pKa = 4.29SFNDD35 pKa = 3.73PVAICLDD42 pKa = 3.45ASRR45 pKa = 11.84FDD47 pKa = 3.57LHH49 pKa = 8.19VSEE52 pKa = 6.09DD53 pKa = 4.03ALQHH57 pKa = 4.62THH59 pKa = 6.62LLYY62 pKa = 8.43MTAIGNDD69 pKa = 3.42PQLAYY74 pKa = 9.79ILEE77 pKa = 4.35NRR79 pKa = 11.84LVTDD83 pKa = 4.44GVGWSSDD90 pKa = 3.07GAFRR94 pKa = 11.84YY95 pKa = 8.82HH96 pKa = 7.1KK97 pKa = 10.64KK98 pKa = 10.54GGRR101 pKa = 11.84CSGDD105 pKa = 2.94NDD107 pKa = 3.61TSLGNVIIMLSITYY121 pKa = 8.74AFCEE125 pKa = 4.27EE126 pKa = 4.06SDD128 pKa = 3.54IPHH131 pKa = 6.82IEE133 pKa = 4.0VTNDD137 pKa = 2.82GDD139 pKa = 3.79DD140 pKa = 3.42QVIMVEE146 pKa = 4.21RR147 pKa = 11.84EE148 pKa = 3.84HH149 pKa = 6.13MHH151 pKa = 5.68TVLQIEE157 pKa = 4.22DD158 pKa = 3.8TFRR161 pKa = 11.84KK162 pKa = 9.75FGFLLKK168 pKa = 10.74VEE170 pKa = 4.4DD171 pKa = 3.93PVYY174 pKa = 9.98EE175 pKa = 3.96LEE177 pKa = 6.09RR178 pKa = 11.84IDD180 pKa = 4.18FCQTRR185 pKa = 11.84PVQLTPDD192 pKa = 3.24VCTMVRR198 pKa = 11.84HH199 pKa = 6.23PRR201 pKa = 11.84LAMTKK206 pKa = 10.27DD207 pKa = 3.25LTTFIPIEE215 pKa = 4.15RR216 pKa = 11.84GKK218 pKa = 10.76LKK220 pKa = 10.85YY221 pKa = 10.34HH222 pKa = 6.45MLTAIGKK229 pKa = 9.25CGSACYY235 pKa = 10.46DD236 pKa = 4.55DD237 pKa = 5.5IPVLRR242 pKa = 11.84AMYY245 pKa = 10.26RR246 pKa = 11.84KK247 pKa = 9.06MEE249 pKa = 4.84IIGTRR254 pKa = 11.84NKK256 pKa = 10.54GNEE259 pKa = 3.97DD260 pKa = 3.38SWWNSTHH267 pKa = 7.24ADD269 pKa = 3.41PAFRR273 pKa = 11.84TLSSKK278 pKa = 10.81AATNVGLTTQCRR290 pKa = 11.84ISFWKK295 pKa = 10.69AFGILPDD302 pKa = 3.57QQEE305 pKa = 4.02ALEE308 pKa = 4.25NEE310 pKa = 3.79WRR312 pKa = 11.84QWEE315 pKa = 4.06PSLDD319 pKa = 3.11RR320 pKa = 11.84VEE322 pKa = 5.0FVHH325 pKa = 6.78SFLGDD330 pKa = 3.57YY331 pKa = 7.89PVCEE335 pKa = 4.23GDD337 pKa = 3.51VV338 pKa = 3.25
Molecular weight: 38.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0P5P8|A0A0A0P5P8_9VIRU ORF4 OS=Beet western yellows virus associated RNA OX=1425367 PE=4 SV=1
MM1 pKa = 7.58SPDD4 pKa = 3.46EE5 pKa = 5.1FLACYY10 pKa = 9.97AGRR13 pKa = 11.84KK14 pKa = 5.92KK15 pKa = 9.25TIYY18 pKa = 10.62AKK20 pKa = 10.65AILSLAATPLNIRR33 pKa = 11.84DD34 pKa = 3.5FRR36 pKa = 11.84VRR38 pKa = 11.84AFIKK42 pKa = 10.47KK43 pKa = 9.88EE44 pKa = 3.64KK45 pKa = 10.62DD46 pKa = 2.99KK47 pKa = 11.24LLSSNSDD54 pKa = 3.05PRR56 pKa = 11.84IIQPRR61 pKa = 11.84HH62 pKa = 4.93PRR64 pKa = 11.84FLVSLGRR71 pKa = 11.84YY72 pKa = 8.38IRR74 pKa = 11.84PLEE77 pKa = 3.92QIVYY81 pKa = 10.3KK82 pKa = 9.49SLRR85 pKa = 11.84DD86 pKa = 3.73CSSDD90 pKa = 3.34SQSQIPPCASKK101 pKa = 11.11ASITSGGGILL111 pKa = 4.47
MM1 pKa = 7.58SPDD4 pKa = 3.46EE5 pKa = 5.1FLACYY10 pKa = 9.97AGRR13 pKa = 11.84KK14 pKa = 5.92KK15 pKa = 9.25TIYY18 pKa = 10.62AKK20 pKa = 10.65AILSLAATPLNIRR33 pKa = 11.84DD34 pKa = 3.5FRR36 pKa = 11.84VRR38 pKa = 11.84AFIKK42 pKa = 10.47KK43 pKa = 9.88EE44 pKa = 3.64KK45 pKa = 10.62DD46 pKa = 2.99KK47 pKa = 11.24LLSSNSDD54 pKa = 3.05PRR56 pKa = 11.84IIQPRR61 pKa = 11.84HH62 pKa = 4.93PRR64 pKa = 11.84FLVSLGRR71 pKa = 11.84YY72 pKa = 8.38IRR74 pKa = 11.84PLEE77 pKa = 3.92QIVYY81 pKa = 10.3KK82 pKa = 9.49SLRR85 pKa = 11.84DD86 pKa = 3.73CSSDD90 pKa = 3.34SQSQIPPCASKK101 pKa = 11.11ASITSGGGILL111 pKa = 4.47
Molecular weight: 12.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
477 |
28 |
338 |
159.0 |
18.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.66 ± 1.008 | 3.145 ± 0.662 |
7.338 ± 0.847 | 5.241 ± 1.628 |
4.612 ± 1.024 | 5.66 ± 0.702 |
2.935 ± 0.957 | 6.289 ± 1.612 |
5.241 ± 1.294 | 8.386 ± 0.893 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.725 ± 0.757 | 3.354 ± 0.661 |
5.031 ± 1.11 | 3.354 ± 0.7 |
7.338 ± 1.587 | 6.918 ± 2.35 |
6.709 ± 2.615 | 5.451 ± 1.326 |
1.468 ± 0.753 | 3.145 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
