
Ntepes virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Ntepes phlebovirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
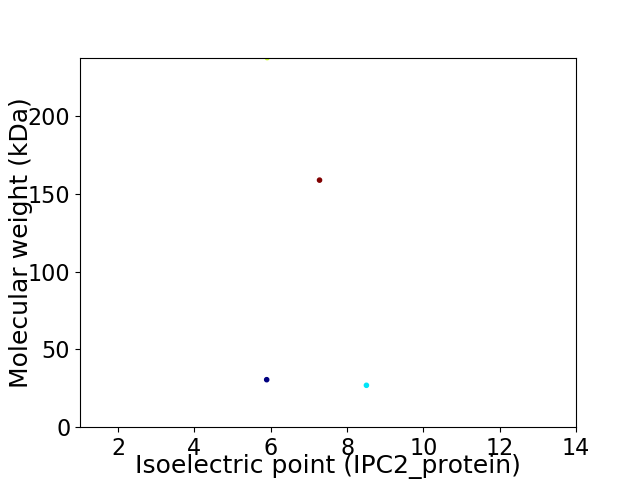
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
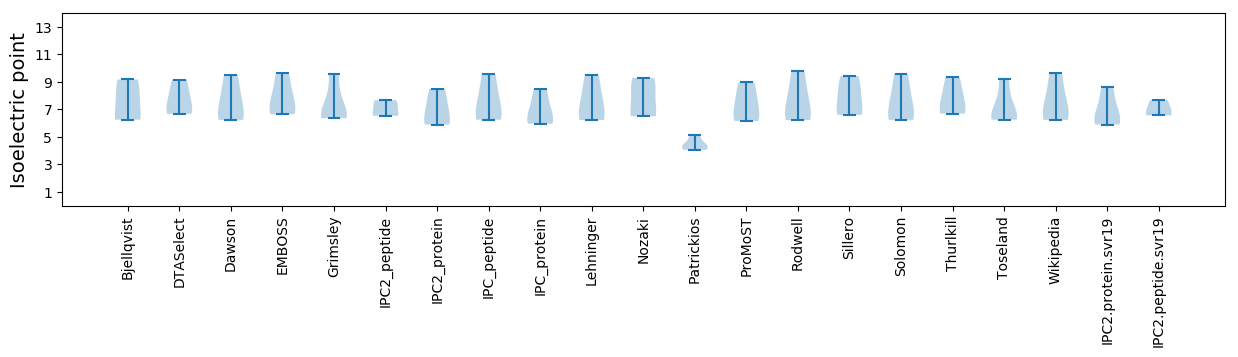
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A499S7X0|A0A499S7X0_9VIRU Replicase OS=Ntepes virus OX=2569589 GN=RdRp PE=4 SV=1
MM1 pKa = 7.81KK2 pKa = 8.03MTTRR6 pKa = 11.84FLYY9 pKa = 10.54DD10 pKa = 3.15RR11 pKa = 11.84LYY13 pKa = 11.2VMQLEE18 pKa = 4.55GQKK21 pKa = 10.35LQAVFLAHH29 pKa = 6.8NNFVPNDD36 pKa = 3.5VSSYY40 pKa = 10.66DD41 pKa = 3.27HH42 pKa = 6.59MEE44 pKa = 4.16VVLSKK49 pKa = 10.55YY50 pKa = 9.82RR51 pKa = 11.84QSFDD55 pKa = 3.34YY56 pKa = 10.87RR57 pKa = 11.84EE58 pKa = 4.21SLSDD62 pKa = 4.12FYY64 pKa = 11.8SQGEE68 pKa = 4.12LPFRR72 pKa = 11.84WGSVMWCSRR81 pKa = 11.84VTGEE85 pKa = 4.53EE86 pKa = 4.15YY87 pKa = 10.83LSFMALMMDD96 pKa = 4.26LVKK99 pKa = 10.17IDD101 pKa = 3.55SSEE104 pKa = 4.15LKK106 pKa = 10.93GNEE109 pKa = 3.91HH110 pKa = 6.34PNIRR114 pKa = 11.84EE115 pKa = 3.96ALSWPTGSPTMGFIKK130 pKa = 10.32LNCLGSTWSLNYY142 pKa = 9.83QKK144 pKa = 11.06SRR146 pKa = 11.84VATLLLRR153 pKa = 11.84AGGSKK158 pKa = 10.55DD159 pKa = 3.31GLEE162 pKa = 3.83EE163 pKa = 5.6AIVKK167 pKa = 6.08THH169 pKa = 6.14KK170 pKa = 10.26KK171 pKa = 9.8ILMEE175 pKa = 4.02SSIRR179 pKa = 11.84GFDD182 pKa = 3.48PKK184 pKa = 10.61HH185 pKa = 6.05FPGLDD190 pKa = 4.1LIRR193 pKa = 11.84EE194 pKa = 4.38VACLQCVRR202 pKa = 11.84LMNASCFDD210 pKa = 4.14SVHH213 pKa = 5.56TAYY216 pKa = 9.9PSRR219 pKa = 11.84LLDD222 pKa = 3.5VLSMHH227 pKa = 7.09RR228 pKa = 11.84STYY231 pKa = 10.04TMISSKK237 pKa = 10.93LLGNRR242 pKa = 11.84KK243 pKa = 5.5WTPVQDD249 pKa = 3.55THH251 pKa = 8.57FDD253 pKa = 3.88EE254 pKa = 6.07PEE256 pKa = 3.54ATYY259 pKa = 11.04DD260 pKa = 3.65FSSDD264 pKa = 3.39SEE266 pKa = 4.28
MM1 pKa = 7.81KK2 pKa = 8.03MTTRR6 pKa = 11.84FLYY9 pKa = 10.54DD10 pKa = 3.15RR11 pKa = 11.84LYY13 pKa = 11.2VMQLEE18 pKa = 4.55GQKK21 pKa = 10.35LQAVFLAHH29 pKa = 6.8NNFVPNDD36 pKa = 3.5VSSYY40 pKa = 10.66DD41 pKa = 3.27HH42 pKa = 6.59MEE44 pKa = 4.16VVLSKK49 pKa = 10.55YY50 pKa = 9.82RR51 pKa = 11.84QSFDD55 pKa = 3.34YY56 pKa = 10.87RR57 pKa = 11.84EE58 pKa = 4.21SLSDD62 pKa = 4.12FYY64 pKa = 11.8SQGEE68 pKa = 4.12LPFRR72 pKa = 11.84WGSVMWCSRR81 pKa = 11.84VTGEE85 pKa = 4.53EE86 pKa = 4.15YY87 pKa = 10.83LSFMALMMDD96 pKa = 4.26LVKK99 pKa = 10.17IDD101 pKa = 3.55SSEE104 pKa = 4.15LKK106 pKa = 10.93GNEE109 pKa = 3.91HH110 pKa = 6.34PNIRR114 pKa = 11.84EE115 pKa = 3.96ALSWPTGSPTMGFIKK130 pKa = 10.32LNCLGSTWSLNYY142 pKa = 9.83QKK144 pKa = 11.06SRR146 pKa = 11.84VATLLLRR153 pKa = 11.84AGGSKK158 pKa = 10.55DD159 pKa = 3.31GLEE162 pKa = 3.83EE163 pKa = 5.6AIVKK167 pKa = 6.08THH169 pKa = 6.14KK170 pKa = 10.26KK171 pKa = 9.8ILMEE175 pKa = 4.02SSIRR179 pKa = 11.84GFDD182 pKa = 3.48PKK184 pKa = 10.61HH185 pKa = 6.05FPGLDD190 pKa = 4.1LIRR193 pKa = 11.84EE194 pKa = 4.38VACLQCVRR202 pKa = 11.84LMNASCFDD210 pKa = 4.14SVHH213 pKa = 5.56TAYY216 pKa = 9.9PSRR219 pKa = 11.84LLDD222 pKa = 3.5VLSMHH227 pKa = 7.09RR228 pKa = 11.84STYY231 pKa = 10.04TMISSKK237 pKa = 10.93LLGNRR242 pKa = 11.84KK243 pKa = 5.5WTPVQDD249 pKa = 3.55THH251 pKa = 8.57FDD253 pKa = 3.88EE254 pKa = 6.07PEE256 pKa = 3.54ATYY259 pKa = 11.04DD260 pKa = 3.65FSSDD264 pKa = 3.39SEE266 pKa = 4.28
Molecular weight: 30.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A499S1A8|A0A499S1A8_9VIRU Glycoprotein OS=Ntepes virus OX=2569589 GN=GPC PE=4 SV=1
MM1 pKa = 7.7SDD3 pKa = 3.29YY4 pKa = 11.22AQIAIEE10 pKa = 4.47FGSSGIDD17 pKa = 3.09RR18 pKa = 11.84DD19 pKa = 5.08LIEE22 pKa = 3.85AWVRR26 pKa = 11.84EE27 pKa = 3.95FEE29 pKa = 4.4YY30 pKa = 10.73QGFDD34 pKa = 3.13PKK36 pKa = 11.2VVVKK40 pKa = 10.49IVTEE44 pKa = 4.23TGEE47 pKa = 4.03GWQNDD52 pKa = 3.92VKK54 pKa = 11.4KK55 pKa = 10.31MIILALTRR63 pKa = 11.84GNKK66 pKa = 7.57PEE68 pKa = 3.97KK69 pKa = 9.99MIAKK73 pKa = 8.73MSAKK77 pKa = 9.62GRR79 pKa = 11.84EE80 pKa = 3.96EE81 pKa = 3.68VTRR84 pKa = 11.84LVKK87 pKa = 10.21KK88 pKa = 10.84YY89 pKa = 10.07KK90 pKa = 10.37LKK92 pKa = 10.78SGNPGRR98 pKa = 11.84NDD100 pKa = 3.09LTLSRR105 pKa = 11.84VAAAFASWTCAAIYY119 pKa = 9.27YY120 pKa = 8.47VQDD123 pKa = 3.73YY124 pKa = 11.05LPVTGSHH131 pKa = 6.29MDD133 pKa = 4.16AISRR137 pKa = 11.84NYY139 pKa = 8.3PRR141 pKa = 11.84AMMHH145 pKa = 6.7PSFAGLIDD153 pKa = 3.67PGMRR157 pKa = 11.84DD158 pKa = 3.18RR159 pKa = 11.84EE160 pKa = 4.29ILVEE164 pKa = 3.96AHH166 pKa = 6.84SLFLIEE172 pKa = 5.02FAQKK176 pKa = 10.53INVNLRR182 pKa = 11.84GKK184 pKa = 10.14SKK186 pKa = 11.46AEE188 pKa = 3.55ILLSFDD194 pKa = 3.42QPLNAAINSNFLTSSQKK211 pKa = 10.78LKK213 pKa = 10.99ALAALGVIDD222 pKa = 4.61EE223 pKa = 4.42HH224 pKa = 6.63QKK226 pKa = 9.98PNRR229 pKa = 11.84DD230 pKa = 3.26VVSAADD236 pKa = 3.89AFRR239 pKa = 11.84KK240 pKa = 8.23MM241 pKa = 3.96
MM1 pKa = 7.7SDD3 pKa = 3.29YY4 pKa = 11.22AQIAIEE10 pKa = 4.47FGSSGIDD17 pKa = 3.09RR18 pKa = 11.84DD19 pKa = 5.08LIEE22 pKa = 3.85AWVRR26 pKa = 11.84EE27 pKa = 3.95FEE29 pKa = 4.4YY30 pKa = 10.73QGFDD34 pKa = 3.13PKK36 pKa = 11.2VVVKK40 pKa = 10.49IVTEE44 pKa = 4.23TGEE47 pKa = 4.03GWQNDD52 pKa = 3.92VKK54 pKa = 11.4KK55 pKa = 10.31MIILALTRR63 pKa = 11.84GNKK66 pKa = 7.57PEE68 pKa = 3.97KK69 pKa = 9.99MIAKK73 pKa = 8.73MSAKK77 pKa = 9.62GRR79 pKa = 11.84EE80 pKa = 3.96EE81 pKa = 3.68VTRR84 pKa = 11.84LVKK87 pKa = 10.21KK88 pKa = 10.84YY89 pKa = 10.07KK90 pKa = 10.37LKK92 pKa = 10.78SGNPGRR98 pKa = 11.84NDD100 pKa = 3.09LTLSRR105 pKa = 11.84VAAAFASWTCAAIYY119 pKa = 9.27YY120 pKa = 8.47VQDD123 pKa = 3.73YY124 pKa = 11.05LPVTGSHH131 pKa = 6.29MDD133 pKa = 4.16AISRR137 pKa = 11.84NYY139 pKa = 8.3PRR141 pKa = 11.84AMMHH145 pKa = 6.7PSFAGLIDD153 pKa = 3.67PGMRR157 pKa = 11.84DD158 pKa = 3.18RR159 pKa = 11.84EE160 pKa = 4.29ILVEE164 pKa = 3.96AHH166 pKa = 6.84SLFLIEE172 pKa = 5.02FAQKK176 pKa = 10.53INVNLRR182 pKa = 11.84GKK184 pKa = 10.14SKK186 pKa = 11.46AEE188 pKa = 3.55ILLSFDD194 pKa = 3.42QPLNAAINSNFLTSSQKK211 pKa = 10.78LKK213 pKa = 10.99ALAALGVIDD222 pKa = 4.61EE223 pKa = 4.42HH224 pKa = 6.63QKK226 pKa = 9.98PNRR229 pKa = 11.84DD230 pKa = 3.26VVSAADD236 pKa = 3.89AFRR239 pKa = 11.84KK240 pKa = 8.23MM241 pKa = 3.96
Molecular weight: 26.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4038 |
241 |
2102 |
1009.5 |
113.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.894 ± 0.722 | 2.526 ± 0.668 |
5.225 ± 0.349 | 7.231 ± 0.364 |
4.383 ± 0.714 | 6.142 ± 0.175 |
2.377 ± 0.19 | 6.662 ± 0.608 |
6.612 ± 0.502 | 8.692 ± 0.404 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.12 ± 0.546 | 3.962 ± 0.288 |
3.888 ± 0.222 | 2.774 ± 0.304 |
5.721 ± 0.213 | 9.039 ± 0.369 |
5.869 ± 0.674 | 6.142 ± 0.153 |
1.189 ± 0.161 | 2.551 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
