
Methanolacinia petrolearia (strain DSM 11571 / OCM 486 / SEBR 4847) (Methanoplanus petrolearius)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanomicrobiaceae; Methanolacinia; Methanolacinia petrolearia
Average proteome isoelectric point is 5.59
Get precalculated fractions of proteins
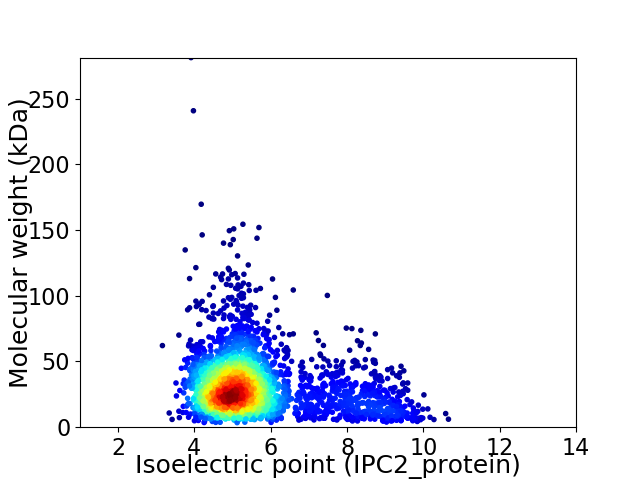
Virtual 2D-PAGE plot for 2779 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|E1RFT3|E1RFT3_METP4 ABC transporter related protein OS=Methanolacinia petrolearia (strain DSM 11571 / OCM 486 / SEBR 4847) OX=679926 GN=Mpet_0311 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.04KK3 pKa = 9.86HH4 pKa = 6.0VIAVLIAVAILSGIFFIAPCFFDD27 pKa = 3.89SSPGSEE33 pKa = 4.03DD34 pKa = 3.1ATLKK38 pKa = 10.68SVQNPDD44 pKa = 2.67TGSIHH49 pKa = 7.08RR50 pKa = 11.84EE51 pKa = 3.73NVSHH55 pKa = 7.27NEE57 pKa = 3.73TTEE60 pKa = 3.84PSGNDD65 pKa = 3.77CEE67 pKa = 4.93DD68 pKa = 3.58CCLVEE73 pKa = 5.51SGDD76 pKa = 3.37RR77 pKa = 11.84SYY79 pKa = 11.98GVMSIDD85 pKa = 4.98DD86 pKa = 3.97EE87 pKa = 4.58YY88 pKa = 11.64KK89 pKa = 10.03EE90 pKa = 4.25EE91 pKa = 4.48KK92 pKa = 10.63EE93 pKa = 3.9EE94 pKa = 4.18TINANPVVSYY104 pKa = 9.04ATEE107 pKa = 4.0NPSVGKK113 pKa = 8.88TDD115 pKa = 4.05FNDD118 pKa = 5.39DD119 pKa = 3.89IPDD122 pKa = 3.61SSGSGEE128 pKa = 4.12NLSGYY133 pKa = 9.62IPYY136 pKa = 10.52FGDD139 pKa = 3.39EE140 pKa = 4.44RR141 pKa = 11.84DD142 pKa = 3.4QGLCGNCWVWTCTEE156 pKa = 3.7IAEE159 pKa = 4.39IEE161 pKa = 4.52YY162 pKa = 10.68AVQTGNKK169 pKa = 8.55EE170 pKa = 4.15RR171 pKa = 11.84FSVQYY176 pKa = 10.71FNSNYY181 pKa = 10.22KK182 pKa = 10.53NGTGSGASASWACCGGNLFSFTGFYY207 pKa = 10.96DD208 pKa = 4.79LNDD211 pKa = 3.69QKK213 pKa = 11.75KK214 pKa = 10.35LIPWNNTKK222 pKa = 10.4ARR224 pKa = 11.84YY225 pKa = 9.13YY226 pKa = 11.0DD227 pKa = 4.33LLIKK231 pKa = 10.8CSDD234 pKa = 3.75GASNMQSALISEE246 pKa = 4.8DD247 pKa = 3.1PSVPFEE253 pKa = 4.14SVAPFTVQTWGISQSVAIDD272 pKa = 3.87TIEE275 pKa = 4.21SYY277 pKa = 10.74IDD279 pKa = 3.09QGRR282 pKa = 11.84ALYY285 pKa = 10.75VSFEE289 pKa = 4.33WVSGEE294 pKa = 5.96GFDD297 pKa = 4.09QWWSSEE303 pKa = 4.21SEE305 pKa = 3.99EE306 pKa = 4.94SVYY309 pKa = 11.25DD310 pKa = 4.17PIGDD314 pKa = 3.92MGNGTPDD321 pKa = 3.12SAHH324 pKa = 6.26AVTLIGYY331 pKa = 9.4NEE333 pKa = 4.34TAWLLHH339 pKa = 5.84NSWGTSSNHH348 pKa = 6.56PDD350 pKa = 2.83GTFWMTKK357 pKa = 10.4DD358 pKa = 3.22FDD360 pKa = 4.71YY361 pKa = 11.53NAGSLLFPSDD371 pKa = 3.66TYY373 pKa = 11.07VNFYY377 pKa = 10.78GFEE380 pKa = 3.97ITGLTEE386 pKa = 3.82TAPDD390 pKa = 4.78LIRR393 pKa = 11.84EE394 pKa = 4.1YY395 pKa = 10.65TYY397 pKa = 11.47GGTNADD403 pKa = 3.26AAYY406 pKa = 9.79GIVSASDD413 pKa = 3.06GGYY416 pKa = 10.18VLAGEE421 pKa = 4.54KK422 pKa = 10.16SSKK425 pKa = 9.76NGNYY429 pKa = 8.78PLNYY433 pKa = 9.46GSSDD437 pKa = 2.55TWMVKK442 pKa = 9.08TNSSGSVEE450 pKa = 3.78WEE452 pKa = 3.63INYY455 pKa = 10.1GGSGDD460 pKa = 3.78DD461 pKa = 3.18TAFAIIKK468 pKa = 9.57TSDD471 pKa = 3.44DD472 pKa = 3.46GFLIAGSTSSNDD484 pKa = 3.06INISGNNGGVDD495 pKa = 3.79ALLLKK500 pKa = 9.84TDD502 pKa = 4.01SNGVLQWLKK511 pKa = 11.02VYY513 pKa = 10.51GGSGDD518 pKa = 4.76DD519 pKa = 3.51SFNDD523 pKa = 3.2IEE525 pKa = 4.44AVSGGYY531 pKa = 9.84LAVGEE536 pKa = 4.53TKK538 pKa = 10.69SSGGDD543 pKa = 3.13VSSNNGGGDD552 pKa = 3.04AWIVKK557 pKa = 9.75VDD559 pKa = 3.53TSGDD563 pKa = 3.7ILWEE567 pKa = 3.86NTYY570 pKa = 10.78GGSADD575 pKa = 3.95EE576 pKa = 4.99SISDD580 pKa = 4.62IILPGDD586 pKa = 3.64GTLLASGSTASSDD599 pKa = 3.82GNVSSNNGGEE609 pKa = 4.15DD610 pKa = 2.7AWIIKK615 pKa = 9.14TGLSGDD621 pKa = 4.26LLWEE625 pKa = 3.9NTYY628 pKa = 10.86GGSGDD633 pKa = 3.78EE634 pKa = 4.0QALGIGVLSDD644 pKa = 3.06GSYY647 pKa = 10.34IVAGSAEE654 pKa = 4.23SSDD657 pKa = 3.77GDD659 pKa = 3.54VSSNNGCDD667 pKa = 3.81DD668 pKa = 3.03VWLFRR673 pKa = 11.84INSSGSLEE681 pKa = 3.91WEE683 pKa = 3.99KK684 pKa = 11.44NYY686 pKa = 10.95GGSGNDD692 pKa = 3.08AAASVLVLDD701 pKa = 4.92DD702 pKa = 6.33DD703 pKa = 5.11YY704 pKa = 11.92ILVCGATGSSDD715 pKa = 3.38GDD717 pKa = 3.62VAGNNGDD724 pKa = 3.6TDD726 pKa = 3.22VWLIKK731 pKa = 9.66TDD733 pKa = 3.43SSGDD737 pKa = 3.76IVWTGCHH744 pKa = 6.25GGQKK748 pKa = 9.94KK749 pKa = 10.4DD750 pKa = 4.02LIYY753 pKa = 10.77EE754 pKa = 4.35LVPSGSVNGFIGGGCTFSSDD774 pKa = 3.45GDD776 pKa = 3.59VSFNNGGGDD785 pKa = 3.3LWIAEE790 pKa = 4.04FGYY793 pKa = 10.0PDD795 pKa = 4.74EE796 pKa = 5.85NPMPDD801 pKa = 3.45PTVTPTQRR809 pKa = 11.84LTVTPTPTEE818 pKa = 4.09VYY820 pKa = 8.98TKK822 pKa = 10.83VSGEE826 pKa = 3.99PSSTSVDD833 pKa = 3.37LQAIIRR839 pKa = 11.84VMSEE843 pKa = 3.26KK844 pKa = 10.57DD845 pKa = 3.28YY846 pKa = 10.29TADD849 pKa = 3.1TGIEE853 pKa = 4.07GGEE856 pKa = 4.1QEE858 pKa = 5.33SDD860 pKa = 3.32GQQTEE865 pKa = 4.2TPPYY869 pKa = 10.07INAQKK874 pKa = 10.81VIGGTGDD881 pKa = 4.07DD882 pKa = 4.07FATSVIAGSSGFVVAGTVDD901 pKa = 3.38SDD903 pKa = 3.52EE904 pKa = 5.13GDD906 pKa = 3.33IEE908 pKa = 4.52SEE910 pKa = 4.0NGGADD915 pKa = 3.06ILLAGFDD922 pKa = 3.89SGLDD926 pKa = 3.79LVWQKK931 pKa = 11.44YY932 pKa = 9.85IGGSGDD938 pKa = 3.48DD939 pKa = 3.55TAVSIVKK946 pKa = 10.46SGDD949 pKa = 3.37DD950 pKa = 3.42SLVIAGTTSSSDD962 pKa = 2.84GDD964 pKa = 3.46IPLLHH969 pKa = 6.87GGSDD973 pKa = 3.15ILLACTDD980 pKa = 3.51GKK982 pKa = 11.62GDD984 pKa = 3.81IKK986 pKa = 10.19WVKK989 pKa = 9.95SAGGSLDD996 pKa = 3.91DD997 pKa = 4.75RR998 pKa = 11.84ASSMIMTDD1006 pKa = 3.21DD1007 pKa = 3.58GNFIVVGEE1015 pKa = 4.33TWSSDD1020 pKa = 2.74GDD1022 pKa = 3.24ISFEE1026 pKa = 3.75NRR1028 pKa = 11.84RR1029 pKa = 11.84NLGEE1033 pKa = 4.28GDD1035 pKa = 3.29LWAVCFDD1042 pKa = 3.67SSGNLVWEE1050 pKa = 4.3RR1051 pKa = 11.84RR1052 pKa = 11.84LGGTGYY1058 pKa = 10.98DD1059 pKa = 3.46SASAVAPAPGGGCTIAGQTRR1079 pKa = 11.84SYY1081 pKa = 11.48DD1082 pKa = 3.55GDD1084 pKa = 3.46PSGNNKK1090 pKa = 10.01HH1091 pKa = 6.25YY1092 pKa = 10.61GEE1094 pKa = 4.06SDD1096 pKa = 2.49IWVIRR1101 pKa = 11.84LDD1103 pKa = 3.45RR1104 pKa = 11.84YY1105 pKa = 10.8GRR1107 pKa = 11.84IIWQKK1112 pKa = 8.28TFGTSGEE1119 pKa = 4.49DD1120 pKa = 3.3GATDD1124 pKa = 3.59IEE1126 pKa = 4.5LSGDD1130 pKa = 3.04GCYY1133 pKa = 10.08IISGYY1138 pKa = 10.1AAAGDD1143 pKa = 3.89DD1144 pKa = 4.42DD1145 pKa = 5.0AAGSYY1150 pKa = 10.19GGSDD1154 pKa = 3.49ALILKK1159 pKa = 9.6IDD1161 pKa = 3.84SSGKK1165 pKa = 9.78LLWKK1169 pKa = 10.19RR1170 pKa = 11.84SYY1172 pKa = 11.2GSSSSDD1178 pKa = 2.97LGIAAEE1184 pKa = 4.44CCSNGDD1190 pKa = 3.51FVIAGVSFNKK1200 pKa = 9.02PGNMSSWGDD1209 pKa = 3.32YY1210 pKa = 10.77DD1211 pKa = 3.3GWLIRR1216 pKa = 11.84LDD1218 pKa = 3.61PSGTVLWEE1226 pKa = 3.8KK1227 pKa = 11.12DD1228 pKa = 3.5FGSSGEE1234 pKa = 4.02DD1235 pKa = 2.45WGYY1238 pKa = 11.17DD1239 pKa = 2.79IDD1241 pKa = 4.03YY1242 pKa = 10.32CAEE1245 pKa = 3.66EE1246 pKa = 4.81RR1247 pKa = 11.84EE1248 pKa = 4.24FIIAGMTTGLKK1259 pKa = 10.01DD1260 pKa = 3.46HH1261 pKa = 6.63QGSSDD1266 pKa = 3.63SDD1268 pKa = 3.87LLLVKK1273 pKa = 10.03IDD1275 pKa = 5.12DD1276 pKa = 4.51GSADD1280 pKa = 3.25
MM1 pKa = 7.5KK2 pKa = 10.04KK3 pKa = 9.86HH4 pKa = 6.0VIAVLIAVAILSGIFFIAPCFFDD27 pKa = 3.89SSPGSEE33 pKa = 4.03DD34 pKa = 3.1ATLKK38 pKa = 10.68SVQNPDD44 pKa = 2.67TGSIHH49 pKa = 7.08RR50 pKa = 11.84EE51 pKa = 3.73NVSHH55 pKa = 7.27NEE57 pKa = 3.73TTEE60 pKa = 3.84PSGNDD65 pKa = 3.77CEE67 pKa = 4.93DD68 pKa = 3.58CCLVEE73 pKa = 5.51SGDD76 pKa = 3.37RR77 pKa = 11.84SYY79 pKa = 11.98GVMSIDD85 pKa = 4.98DD86 pKa = 3.97EE87 pKa = 4.58YY88 pKa = 11.64KK89 pKa = 10.03EE90 pKa = 4.25EE91 pKa = 4.48KK92 pKa = 10.63EE93 pKa = 3.9EE94 pKa = 4.18TINANPVVSYY104 pKa = 9.04ATEE107 pKa = 4.0NPSVGKK113 pKa = 8.88TDD115 pKa = 4.05FNDD118 pKa = 5.39DD119 pKa = 3.89IPDD122 pKa = 3.61SSGSGEE128 pKa = 4.12NLSGYY133 pKa = 9.62IPYY136 pKa = 10.52FGDD139 pKa = 3.39EE140 pKa = 4.44RR141 pKa = 11.84DD142 pKa = 3.4QGLCGNCWVWTCTEE156 pKa = 3.7IAEE159 pKa = 4.39IEE161 pKa = 4.52YY162 pKa = 10.68AVQTGNKK169 pKa = 8.55EE170 pKa = 4.15RR171 pKa = 11.84FSVQYY176 pKa = 10.71FNSNYY181 pKa = 10.22KK182 pKa = 10.53NGTGSGASASWACCGGNLFSFTGFYY207 pKa = 10.96DD208 pKa = 4.79LNDD211 pKa = 3.69QKK213 pKa = 11.75KK214 pKa = 10.35LIPWNNTKK222 pKa = 10.4ARR224 pKa = 11.84YY225 pKa = 9.13YY226 pKa = 11.0DD227 pKa = 4.33LLIKK231 pKa = 10.8CSDD234 pKa = 3.75GASNMQSALISEE246 pKa = 4.8DD247 pKa = 3.1PSVPFEE253 pKa = 4.14SVAPFTVQTWGISQSVAIDD272 pKa = 3.87TIEE275 pKa = 4.21SYY277 pKa = 10.74IDD279 pKa = 3.09QGRR282 pKa = 11.84ALYY285 pKa = 10.75VSFEE289 pKa = 4.33WVSGEE294 pKa = 5.96GFDD297 pKa = 4.09QWWSSEE303 pKa = 4.21SEE305 pKa = 3.99EE306 pKa = 4.94SVYY309 pKa = 11.25DD310 pKa = 4.17PIGDD314 pKa = 3.92MGNGTPDD321 pKa = 3.12SAHH324 pKa = 6.26AVTLIGYY331 pKa = 9.4NEE333 pKa = 4.34TAWLLHH339 pKa = 5.84NSWGTSSNHH348 pKa = 6.56PDD350 pKa = 2.83GTFWMTKK357 pKa = 10.4DD358 pKa = 3.22FDD360 pKa = 4.71YY361 pKa = 11.53NAGSLLFPSDD371 pKa = 3.66TYY373 pKa = 11.07VNFYY377 pKa = 10.78GFEE380 pKa = 3.97ITGLTEE386 pKa = 3.82TAPDD390 pKa = 4.78LIRR393 pKa = 11.84EE394 pKa = 4.1YY395 pKa = 10.65TYY397 pKa = 11.47GGTNADD403 pKa = 3.26AAYY406 pKa = 9.79GIVSASDD413 pKa = 3.06GGYY416 pKa = 10.18VLAGEE421 pKa = 4.54KK422 pKa = 10.16SSKK425 pKa = 9.76NGNYY429 pKa = 8.78PLNYY433 pKa = 9.46GSSDD437 pKa = 2.55TWMVKK442 pKa = 9.08TNSSGSVEE450 pKa = 3.78WEE452 pKa = 3.63INYY455 pKa = 10.1GGSGDD460 pKa = 3.78DD461 pKa = 3.18TAFAIIKK468 pKa = 9.57TSDD471 pKa = 3.44DD472 pKa = 3.46GFLIAGSTSSNDD484 pKa = 3.06INISGNNGGVDD495 pKa = 3.79ALLLKK500 pKa = 9.84TDD502 pKa = 4.01SNGVLQWLKK511 pKa = 11.02VYY513 pKa = 10.51GGSGDD518 pKa = 4.76DD519 pKa = 3.51SFNDD523 pKa = 3.2IEE525 pKa = 4.44AVSGGYY531 pKa = 9.84LAVGEE536 pKa = 4.53TKK538 pKa = 10.69SSGGDD543 pKa = 3.13VSSNNGGGDD552 pKa = 3.04AWIVKK557 pKa = 9.75VDD559 pKa = 3.53TSGDD563 pKa = 3.7ILWEE567 pKa = 3.86NTYY570 pKa = 10.78GGSADD575 pKa = 3.95EE576 pKa = 4.99SISDD580 pKa = 4.62IILPGDD586 pKa = 3.64GTLLASGSTASSDD599 pKa = 3.82GNVSSNNGGEE609 pKa = 4.15DD610 pKa = 2.7AWIIKK615 pKa = 9.14TGLSGDD621 pKa = 4.26LLWEE625 pKa = 3.9NTYY628 pKa = 10.86GGSGDD633 pKa = 3.78EE634 pKa = 4.0QALGIGVLSDD644 pKa = 3.06GSYY647 pKa = 10.34IVAGSAEE654 pKa = 4.23SSDD657 pKa = 3.77GDD659 pKa = 3.54VSSNNGCDD667 pKa = 3.81DD668 pKa = 3.03VWLFRR673 pKa = 11.84INSSGSLEE681 pKa = 3.91WEE683 pKa = 3.99KK684 pKa = 11.44NYY686 pKa = 10.95GGSGNDD692 pKa = 3.08AAASVLVLDD701 pKa = 4.92DD702 pKa = 6.33DD703 pKa = 5.11YY704 pKa = 11.92ILVCGATGSSDD715 pKa = 3.38GDD717 pKa = 3.62VAGNNGDD724 pKa = 3.6TDD726 pKa = 3.22VWLIKK731 pKa = 9.66TDD733 pKa = 3.43SSGDD737 pKa = 3.76IVWTGCHH744 pKa = 6.25GGQKK748 pKa = 9.94KK749 pKa = 10.4DD750 pKa = 4.02LIYY753 pKa = 10.77EE754 pKa = 4.35LVPSGSVNGFIGGGCTFSSDD774 pKa = 3.45GDD776 pKa = 3.59VSFNNGGGDD785 pKa = 3.3LWIAEE790 pKa = 4.04FGYY793 pKa = 10.0PDD795 pKa = 4.74EE796 pKa = 5.85NPMPDD801 pKa = 3.45PTVTPTQRR809 pKa = 11.84LTVTPTPTEE818 pKa = 4.09VYY820 pKa = 8.98TKK822 pKa = 10.83VSGEE826 pKa = 3.99PSSTSVDD833 pKa = 3.37LQAIIRR839 pKa = 11.84VMSEE843 pKa = 3.26KK844 pKa = 10.57DD845 pKa = 3.28YY846 pKa = 10.29TADD849 pKa = 3.1TGIEE853 pKa = 4.07GGEE856 pKa = 4.1QEE858 pKa = 5.33SDD860 pKa = 3.32GQQTEE865 pKa = 4.2TPPYY869 pKa = 10.07INAQKK874 pKa = 10.81VIGGTGDD881 pKa = 4.07DD882 pKa = 4.07FATSVIAGSSGFVVAGTVDD901 pKa = 3.38SDD903 pKa = 3.52EE904 pKa = 5.13GDD906 pKa = 3.33IEE908 pKa = 4.52SEE910 pKa = 4.0NGGADD915 pKa = 3.06ILLAGFDD922 pKa = 3.89SGLDD926 pKa = 3.79LVWQKK931 pKa = 11.44YY932 pKa = 9.85IGGSGDD938 pKa = 3.48DD939 pKa = 3.55TAVSIVKK946 pKa = 10.46SGDD949 pKa = 3.37DD950 pKa = 3.42SLVIAGTTSSSDD962 pKa = 2.84GDD964 pKa = 3.46IPLLHH969 pKa = 6.87GGSDD973 pKa = 3.15ILLACTDD980 pKa = 3.51GKK982 pKa = 11.62GDD984 pKa = 3.81IKK986 pKa = 10.19WVKK989 pKa = 9.95SAGGSLDD996 pKa = 3.91DD997 pKa = 4.75RR998 pKa = 11.84ASSMIMTDD1006 pKa = 3.21DD1007 pKa = 3.58GNFIVVGEE1015 pKa = 4.33TWSSDD1020 pKa = 2.74GDD1022 pKa = 3.24ISFEE1026 pKa = 3.75NRR1028 pKa = 11.84RR1029 pKa = 11.84NLGEE1033 pKa = 4.28GDD1035 pKa = 3.29LWAVCFDD1042 pKa = 3.67SSGNLVWEE1050 pKa = 4.3RR1051 pKa = 11.84RR1052 pKa = 11.84LGGTGYY1058 pKa = 10.98DD1059 pKa = 3.46SASAVAPAPGGGCTIAGQTRR1079 pKa = 11.84SYY1081 pKa = 11.48DD1082 pKa = 3.55GDD1084 pKa = 3.46PSGNNKK1090 pKa = 10.01HH1091 pKa = 6.25YY1092 pKa = 10.61GEE1094 pKa = 4.06SDD1096 pKa = 2.49IWVIRR1101 pKa = 11.84LDD1103 pKa = 3.45RR1104 pKa = 11.84YY1105 pKa = 10.8GRR1107 pKa = 11.84IIWQKK1112 pKa = 8.28TFGTSGEE1119 pKa = 4.49DD1120 pKa = 3.3GATDD1124 pKa = 3.59IEE1126 pKa = 4.5LSGDD1130 pKa = 3.04GCYY1133 pKa = 10.08IISGYY1138 pKa = 10.1AAAGDD1143 pKa = 3.89DD1144 pKa = 4.42DD1145 pKa = 5.0AAGSYY1150 pKa = 10.19GGSDD1154 pKa = 3.49ALILKK1159 pKa = 9.6IDD1161 pKa = 3.84SSGKK1165 pKa = 9.78LLWKK1169 pKa = 10.19RR1170 pKa = 11.84SYY1172 pKa = 11.2GSSSSDD1178 pKa = 2.97LGIAAEE1184 pKa = 4.44CCSNGDD1190 pKa = 3.51FVIAGVSFNKK1200 pKa = 9.02PGNMSSWGDD1209 pKa = 3.32YY1210 pKa = 10.77DD1211 pKa = 3.3GWLIRR1216 pKa = 11.84LDD1218 pKa = 3.61PSGTVLWEE1226 pKa = 3.8KK1227 pKa = 11.12DD1228 pKa = 3.5FGSSGEE1234 pKa = 4.02DD1235 pKa = 2.45WGYY1238 pKa = 11.17DD1239 pKa = 2.79IDD1241 pKa = 4.03YY1242 pKa = 10.32CAEE1245 pKa = 3.66EE1246 pKa = 4.81RR1247 pKa = 11.84EE1248 pKa = 4.24FIIAGMTTGLKK1259 pKa = 10.01DD1260 pKa = 3.46HH1261 pKa = 6.63QGSSDD1266 pKa = 3.63SDD1268 pKa = 3.87LLLVKK1273 pKa = 10.03IDD1275 pKa = 5.12DD1276 pKa = 4.51GSADD1280 pKa = 3.25
Molecular weight: 134.86 kDa
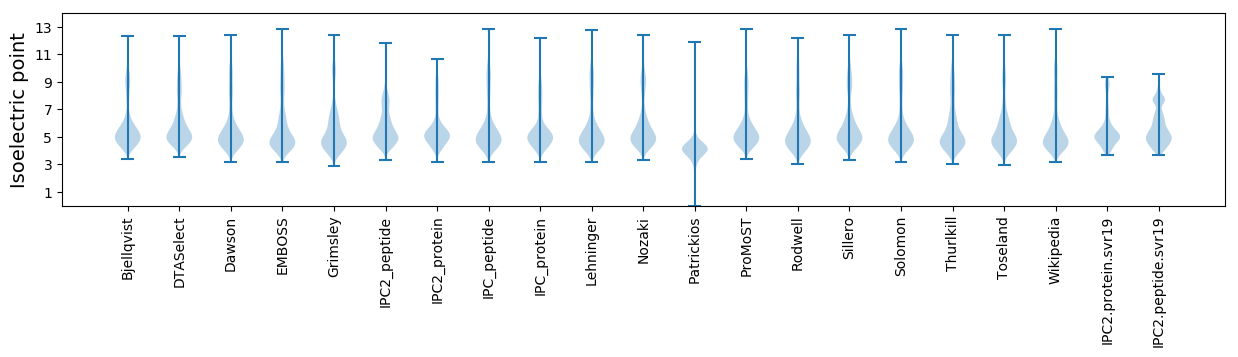
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1RIQ8|E1RIQ8_METP4 Putative nickel insertion protein OS=Methanolacinia petrolearia (strain DSM 11571 / OCM 486 / SEBR 4847) OX=679926 GN=Mpet_1898 PE=3 SV=1
MM1 pKa = 7.88APRR4 pKa = 11.84RR5 pKa = 11.84NKK7 pKa = 8.6NTEE10 pKa = 3.62RR11 pKa = 11.84KK12 pKa = 8.94IAEE15 pKa = 3.93EE16 pKa = 4.03RR17 pKa = 11.84IEE19 pKa = 4.12RR20 pKa = 11.84LFEE23 pKa = 4.12AAHH26 pKa = 5.34TVYY29 pKa = 10.61EE30 pKa = 4.57KK31 pKa = 11.05DD32 pKa = 3.21PSLSGRR38 pKa = 11.84YY39 pKa = 9.05VILAIKK45 pKa = 9.99IAMKK49 pKa = 10.54YY50 pKa = 9.04RR51 pKa = 11.84VKK53 pKa = 10.55IPDD56 pKa = 3.73RR57 pKa = 11.84YY58 pKa = 8.6RR59 pKa = 11.84HH60 pKa = 5.5SYY62 pKa = 9.16CRR64 pKa = 11.84KK65 pKa = 8.18CFSFFSPGNNTRR77 pKa = 11.84TRR79 pKa = 11.84INSGKK84 pKa = 8.64VTLTCTNCGYY94 pKa = 10.28IRR96 pKa = 11.84RR97 pKa = 11.84YY98 pKa = 8.57PIKK101 pKa = 10.15EE102 pKa = 3.69RR103 pKa = 11.84RR104 pKa = 3.5
MM1 pKa = 7.88APRR4 pKa = 11.84RR5 pKa = 11.84NKK7 pKa = 8.6NTEE10 pKa = 3.62RR11 pKa = 11.84KK12 pKa = 8.94IAEE15 pKa = 3.93EE16 pKa = 4.03RR17 pKa = 11.84IEE19 pKa = 4.12RR20 pKa = 11.84LFEE23 pKa = 4.12AAHH26 pKa = 5.34TVYY29 pKa = 10.61EE30 pKa = 4.57KK31 pKa = 11.05DD32 pKa = 3.21PSLSGRR38 pKa = 11.84YY39 pKa = 9.05VILAIKK45 pKa = 9.99IAMKK49 pKa = 10.54YY50 pKa = 9.04RR51 pKa = 11.84VKK53 pKa = 10.55IPDD56 pKa = 3.73RR57 pKa = 11.84YY58 pKa = 8.6RR59 pKa = 11.84HH60 pKa = 5.5SYY62 pKa = 9.16CRR64 pKa = 11.84KK65 pKa = 8.18CFSFFSPGNNTRR77 pKa = 11.84TRR79 pKa = 11.84INSGKK84 pKa = 8.64VTLTCTNCGYY94 pKa = 10.28IRR96 pKa = 11.84RR97 pKa = 11.84YY98 pKa = 8.57PIKK101 pKa = 10.15EE102 pKa = 3.69RR103 pKa = 11.84RR104 pKa = 3.5
Molecular weight: 12.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
826422 |
30 |
2645 |
297.4 |
32.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.241 ± 0.048 | 1.349 ± 0.027 |
5.984 ± 0.039 | 7.475 ± 0.052 |
4.258 ± 0.034 | 7.901 ± 0.045 |
1.549 ± 0.02 | 8.424 ± 0.05 |
5.724 ± 0.048 | 8.67 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.714 ± 0.024 | 4.081 ± 0.036 |
4.122 ± 0.028 | 2.148 ± 0.023 |
4.524 ± 0.042 | 6.95 ± 0.052 |
5.318 ± 0.047 | 7.007 ± 0.041 |
0.95 ± 0.019 | 3.609 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
