
Asystasia mosaic Madagascar virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.22
Get precalculated fractions of proteins
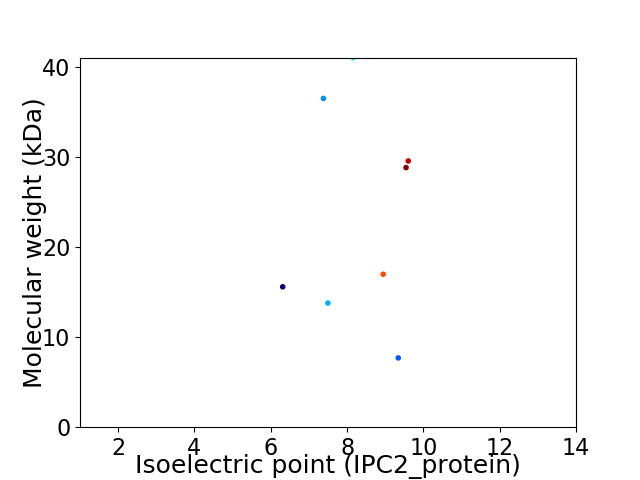
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5BIG2|A0A0C5BIG2_9GEMI Protein V2 OS=Asystasia mosaic Madagascar virus OX=1611435 GN=AV2 PE=3 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84PSSQSPTLSTLVPIKK17 pKa = 10.36VVHH20 pKa = 6.06RR21 pKa = 11.84QAKK24 pKa = 9.65KK25 pKa = 9.27KK26 pKa = 8.57RR27 pKa = 11.84QRR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 10.5KK32 pKa = 9.44IDD34 pKa = 3.66LGCGCSVLIHH44 pKa = 6.26PLCNNYY50 pKa = 9.5GFSHH54 pKa = 7.02RR55 pKa = 11.84GIHH58 pKa = 6.0HH59 pKa = 6.91CSSSAEE65 pKa = 4.0WNILLGTEE73 pKa = 4.38EE74 pKa = 4.81SNIHH78 pKa = 6.14NDD80 pKa = 3.09NRR82 pKa = 11.84THH84 pKa = 6.55RR85 pKa = 11.84QTIQHH90 pKa = 5.93EE91 pKa = 4.4PRR93 pKa = 11.84HH94 pKa = 5.46TEE96 pKa = 3.62HH97 pKa = 7.35PNQVQPQPEE106 pKa = 4.1EE107 pKa = 4.19GFRR110 pKa = 11.84DD111 pKa = 3.55SQMLFEE117 pKa = 4.66LQGLDD122 pKa = 3.71EE123 pKa = 5.12IEE125 pKa = 4.39EE126 pKa = 4.07FDD128 pKa = 3.7ISVLEE133 pKa = 4.38GFF135 pKa = 4.64
MM1 pKa = 7.68RR2 pKa = 11.84PSSQSPTLSTLVPIKK17 pKa = 10.36VVHH20 pKa = 6.06RR21 pKa = 11.84QAKK24 pKa = 9.65KK25 pKa = 9.27KK26 pKa = 8.57RR27 pKa = 11.84QRR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 10.5KK32 pKa = 9.44IDD34 pKa = 3.66LGCGCSVLIHH44 pKa = 6.26PLCNNYY50 pKa = 9.5GFSHH54 pKa = 7.02RR55 pKa = 11.84GIHH58 pKa = 6.0HH59 pKa = 6.91CSSSAEE65 pKa = 4.0WNILLGTEE73 pKa = 4.38EE74 pKa = 4.81SNIHH78 pKa = 6.14NDD80 pKa = 3.09NRR82 pKa = 11.84THH84 pKa = 6.55RR85 pKa = 11.84QTIQHH90 pKa = 5.93EE91 pKa = 4.4PRR93 pKa = 11.84HH94 pKa = 5.46TEE96 pKa = 3.62HH97 pKa = 7.35PNQVQPQPEE106 pKa = 4.1EE107 pKa = 4.19GFRR110 pKa = 11.84DD111 pKa = 3.55SQMLFEE117 pKa = 4.66LQGLDD122 pKa = 3.71EE123 pKa = 5.12IEE125 pKa = 4.39EE126 pKa = 4.07FDD128 pKa = 3.7ISVLEE133 pKa = 4.38GFF135 pKa = 4.64
Molecular weight: 15.57 kDa
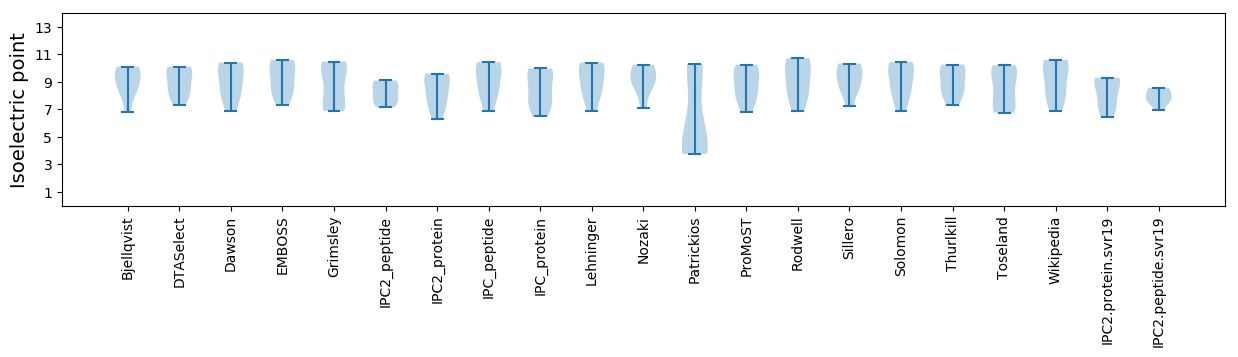
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5B4W5|A0A0C5B4W5_9GEMI Movement protein BC1 OS=Asystasia mosaic Madagascar virus OX=1611435 GN=BC1 PE=3 SV=1
MM1 pKa = 7.7SKK3 pKa = 10.44RR4 pKa = 11.84PGDD7 pKa = 3.89IIISTPASKK16 pKa = 10.49ARR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFDD24 pKa = 3.37SPRR27 pKa = 11.84VAASSVRR34 pKa = 11.84ITRR37 pKa = 11.84RR38 pKa = 11.84QAWVNRR44 pKa = 11.84PMYY47 pKa = 9.72RR48 pKa = 11.84KK49 pKa = 9.29PRR51 pKa = 11.84MYY53 pKa = 10.74RR54 pKa = 11.84MWRR57 pKa = 11.84SYY59 pKa = 10.78DD60 pKa = 3.44VPKK63 pKa = 10.87GCEE66 pKa = 4.43GPCKK70 pKa = 9.67VQSYY74 pKa = 8.13DD75 pKa = 3.16QRR77 pKa = 11.84DD78 pKa = 3.85DD79 pKa = 3.68VKK81 pKa = 10.43HH82 pKa = 6.0VGVVRR87 pKa = 11.84CISDD91 pKa = 3.47VTKK94 pKa = 11.06GPGLTHH100 pKa = 6.06RR101 pKa = 11.84TGKK104 pKa = 10.29RR105 pKa = 11.84FTVKK109 pKa = 9.99SVYY112 pKa = 9.87ICGKK116 pKa = 8.99IWMDD120 pKa = 4.13EE121 pKa = 4.13NIKK124 pKa = 10.06KK125 pKa = 9.97QNHH128 pKa = 4.76TNHH131 pKa = 5.66VLFFLTRR138 pKa = 11.84DD139 pKa = 3.28RR140 pKa = 11.84RR141 pKa = 11.84PYY143 pKa = 9.81GQAPMDD149 pKa = 4.27FGQVFNMFDD158 pKa = 3.99NEE160 pKa = 4.11PSTATVKK167 pKa = 10.85NDD169 pKa = 2.82LRR171 pKa = 11.84DD172 pKa = 3.34RR173 pKa = 11.84FQVLRR178 pKa = 11.84RR179 pKa = 11.84FYY181 pKa = 9.15ATVVGGPSGMKK192 pKa = 9.48EE193 pKa = 3.66QVLVRR198 pKa = 11.84RR199 pKa = 11.84FFKK202 pKa = 11.07LNAQIVYY209 pKa = 9.71NHH211 pKa = 6.32QDD213 pKa = 2.94GAKK216 pKa = 10.03YY217 pKa = 10.3EE218 pKa = 4.13NHH220 pKa = 6.44TEE222 pKa = 3.99NALLLYY228 pKa = 7.29MACTHH233 pKa = 7.07ASNPVYY239 pKa = 10.72ASLKK243 pKa = 8.47VRR245 pKa = 11.84IYY247 pKa = 10.71FYY249 pKa = 11.23DD250 pKa = 3.47SVSNN254 pKa = 3.9
MM1 pKa = 7.7SKK3 pKa = 10.44RR4 pKa = 11.84PGDD7 pKa = 3.89IIISTPASKK16 pKa = 10.49ARR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFDD24 pKa = 3.37SPRR27 pKa = 11.84VAASSVRR34 pKa = 11.84ITRR37 pKa = 11.84RR38 pKa = 11.84QAWVNRR44 pKa = 11.84PMYY47 pKa = 9.72RR48 pKa = 11.84KK49 pKa = 9.29PRR51 pKa = 11.84MYY53 pKa = 10.74RR54 pKa = 11.84MWRR57 pKa = 11.84SYY59 pKa = 10.78DD60 pKa = 3.44VPKK63 pKa = 10.87GCEE66 pKa = 4.43GPCKK70 pKa = 9.67VQSYY74 pKa = 8.13DD75 pKa = 3.16QRR77 pKa = 11.84DD78 pKa = 3.85DD79 pKa = 3.68VKK81 pKa = 10.43HH82 pKa = 6.0VGVVRR87 pKa = 11.84CISDD91 pKa = 3.47VTKK94 pKa = 11.06GPGLTHH100 pKa = 6.06RR101 pKa = 11.84TGKK104 pKa = 10.29RR105 pKa = 11.84FTVKK109 pKa = 9.99SVYY112 pKa = 9.87ICGKK116 pKa = 8.99IWMDD120 pKa = 4.13EE121 pKa = 4.13NIKK124 pKa = 10.06KK125 pKa = 9.97QNHH128 pKa = 4.76TNHH131 pKa = 5.66VLFFLTRR138 pKa = 11.84DD139 pKa = 3.28RR140 pKa = 11.84RR141 pKa = 11.84PYY143 pKa = 9.81GQAPMDD149 pKa = 4.27FGQVFNMFDD158 pKa = 3.99NEE160 pKa = 4.11PSTATVKK167 pKa = 10.85NDD169 pKa = 2.82LRR171 pKa = 11.84DD172 pKa = 3.34RR173 pKa = 11.84FQVLRR178 pKa = 11.84RR179 pKa = 11.84FYY181 pKa = 9.15ATVVGGPSGMKK192 pKa = 9.48EE193 pKa = 3.66QVLVRR198 pKa = 11.84RR199 pKa = 11.84FFKK202 pKa = 11.07LNAQIVYY209 pKa = 9.71NHH211 pKa = 6.32QDD213 pKa = 2.94GAKK216 pKa = 10.03YY217 pKa = 10.3EE218 pKa = 4.13NHH220 pKa = 6.44TEE222 pKa = 3.99NALLLYY228 pKa = 7.29MACTHH233 pKa = 7.07ASNPVYY239 pKa = 10.72ASLKK243 pKa = 8.47VRR245 pKa = 11.84IYY247 pKa = 10.71FYY249 pKa = 11.23DD250 pKa = 3.47SVSNN254 pKa = 3.9
Molecular weight: 29.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1658 |
65 |
362 |
207.3 |
23.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.187 ± 0.76 | 1.99 ± 0.314 |
4.463 ± 0.279 | 4.885 ± 0.822 |
4.704 ± 0.342 | 5.006 ± 0.284 |
3.257 ± 0.425 | 6.333 ± 0.826 |
5.669 ± 0.503 | 6.574 ± 0.447 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.473 ± 0.275 | 5.971 ± 0.55 |
5.127 ± 0.546 | 4.644 ± 0.496 |
7.419 ± 0.848 | 9.771 ± 1.319 |
5.428 ± 0.497 | 6.031 ± 0.918 |
1.086 ± 0.085 | 3.981 ± 0.45 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
