
Mongoose feces-associated gemycircularvirus b
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus monas1; Mongoose associated gemykibivirus 1
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
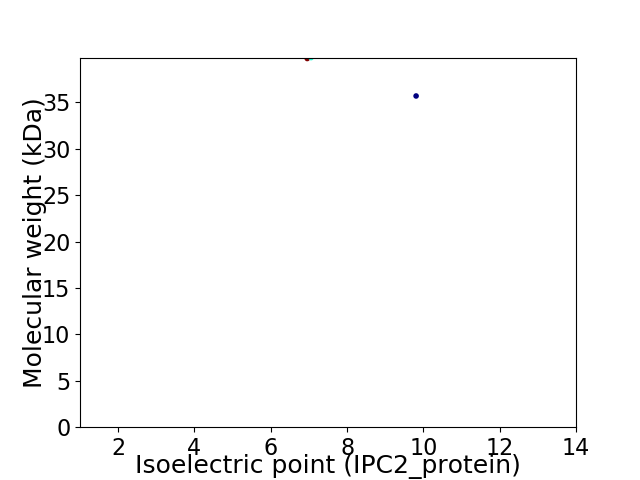
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0E3M1X5|A0A0E3M1X5_9VIRU Rep OS=Mongoose feces-associated gemycircularvirus b OX=1634486 PE=3 SV=1
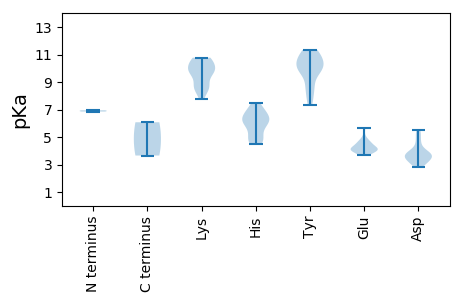
MM1 pKa = 6.85VQYY4 pKa = 10.78NVNNWTIRR12 pKa = 11.84PFRR15 pKa = 11.84KK16 pKa = 9.8LMSSFRR22 pKa = 11.84FQARR26 pKa = 11.84YY27 pKa = 10.11GLFTYY32 pKa = 7.67SQCAQLRR39 pKa = 11.84PDD41 pKa = 5.5AISQHH46 pKa = 5.64YY47 pKa = 8.63EE48 pKa = 3.68AFGADD53 pKa = 3.8YY54 pKa = 10.53IIGRR58 pKa = 11.84EE59 pKa = 3.94THH61 pKa = 6.42SDD63 pKa = 3.12GGTHH67 pKa = 4.98YY68 pKa = 10.73HH69 pKa = 5.75VFVDD73 pKa = 4.7FRR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.91YY78 pKa = 8.97RR79 pKa = 11.84TRR81 pKa = 11.84DD82 pKa = 3.11CRR84 pKa = 11.84KK85 pKa = 8.28FDD87 pKa = 3.69VEE89 pKa = 4.38GFHH92 pKa = 6.93PNIVPSLRR100 pKa = 11.84NPAGGWDD107 pKa = 4.0YY108 pKa = 11.02ATKK111 pKa = 10.74DD112 pKa = 3.63DD113 pKa = 5.46DD114 pKa = 3.76ICGGRR119 pKa = 11.84LRR121 pKa = 11.84RR122 pKa = 11.84PEE124 pKa = 4.06STEE127 pKa = 3.84GNQSGRR133 pKa = 11.84QNAGWDD139 pKa = 3.65EE140 pKa = 4.03LRR142 pKa = 11.84SAEE145 pKa = 4.15TRR147 pKa = 11.84DD148 pKa = 3.4DD149 pKa = 3.8FFRR152 pKa = 11.84IASEE156 pKa = 4.06HH157 pKa = 6.19LFSHH161 pKa = 6.51LVKK164 pKa = 10.53SFHH167 pKa = 6.75SFSAYY172 pKa = 10.39ADD174 pKa = 2.9WKK176 pKa = 9.89YY177 pKa = 10.85RR178 pKa = 11.84VDD180 pKa = 3.73RR181 pKa = 11.84SPYY184 pKa = 7.31QHH186 pKa = 6.44NPEE189 pKa = 4.01FHH191 pKa = 7.18FDD193 pKa = 3.56PGKK196 pKa = 10.77LGTLRR201 pKa = 11.84EE202 pKa = 3.77WHH204 pKa = 5.96GRR206 pKa = 11.84YY207 pKa = 9.75LSDD210 pKa = 4.41HH211 pKa = 6.32IVGKK215 pKa = 9.7GIRR218 pKa = 11.84GMSLLAWGRR227 pKa = 11.84SKK229 pKa = 9.78TGKK232 pKa = 8.59TMWARR237 pKa = 11.84SLGKK241 pKa = 9.22HH242 pKa = 5.95AYY244 pKa = 9.85FGGLFSLDD252 pKa = 2.85EE253 pKa = 4.24WEE255 pKa = 4.07QDD257 pKa = 3.12EE258 pKa = 4.69SVDD261 pKa = 3.64YY262 pKa = 11.3AVFDD266 pKa = 5.29DD267 pKa = 3.86IQGGFKK273 pKa = 10.47FFPAYY278 pKa = 9.75KK279 pKa = 9.8SWLGQQTEE287 pKa = 4.45FHH289 pKa = 6.2CTDD292 pKa = 3.53RR293 pKa = 11.84YY294 pKa = 10.06RR295 pKa = 11.84KK296 pKa = 9.4KK297 pKa = 10.77KK298 pKa = 8.75HH299 pKa = 4.86VQWGKK304 pKa = 8.63PCIWLMNEE312 pKa = 4.66DD313 pKa = 5.19PYY315 pKa = 11.33QQDD318 pKa = 3.06VDD320 pKa = 4.11IDD322 pKa = 3.65WLEE325 pKa = 4.12ANCLIVNVDD334 pKa = 3.02NSLFF338 pKa = 3.65
MM1 pKa = 6.85VQYY4 pKa = 10.78NVNNWTIRR12 pKa = 11.84PFRR15 pKa = 11.84KK16 pKa = 9.8LMSSFRR22 pKa = 11.84FQARR26 pKa = 11.84YY27 pKa = 10.11GLFTYY32 pKa = 7.67SQCAQLRR39 pKa = 11.84PDD41 pKa = 5.5AISQHH46 pKa = 5.64YY47 pKa = 8.63EE48 pKa = 3.68AFGADD53 pKa = 3.8YY54 pKa = 10.53IIGRR58 pKa = 11.84EE59 pKa = 3.94THH61 pKa = 6.42SDD63 pKa = 3.12GGTHH67 pKa = 4.98YY68 pKa = 10.73HH69 pKa = 5.75VFVDD73 pKa = 4.7FRR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.91YY78 pKa = 8.97RR79 pKa = 11.84TRR81 pKa = 11.84DD82 pKa = 3.11CRR84 pKa = 11.84KK85 pKa = 8.28FDD87 pKa = 3.69VEE89 pKa = 4.38GFHH92 pKa = 6.93PNIVPSLRR100 pKa = 11.84NPAGGWDD107 pKa = 4.0YY108 pKa = 11.02ATKK111 pKa = 10.74DD112 pKa = 3.63DD113 pKa = 5.46DD114 pKa = 3.76ICGGRR119 pKa = 11.84LRR121 pKa = 11.84RR122 pKa = 11.84PEE124 pKa = 4.06STEE127 pKa = 3.84GNQSGRR133 pKa = 11.84QNAGWDD139 pKa = 3.65EE140 pKa = 4.03LRR142 pKa = 11.84SAEE145 pKa = 4.15TRR147 pKa = 11.84DD148 pKa = 3.4DD149 pKa = 3.8FFRR152 pKa = 11.84IASEE156 pKa = 4.06HH157 pKa = 6.19LFSHH161 pKa = 6.51LVKK164 pKa = 10.53SFHH167 pKa = 6.75SFSAYY172 pKa = 10.39ADD174 pKa = 2.9WKK176 pKa = 9.89YY177 pKa = 10.85RR178 pKa = 11.84VDD180 pKa = 3.73RR181 pKa = 11.84SPYY184 pKa = 7.31QHH186 pKa = 6.44NPEE189 pKa = 4.01FHH191 pKa = 7.18FDD193 pKa = 3.56PGKK196 pKa = 10.77LGTLRR201 pKa = 11.84EE202 pKa = 3.77WHH204 pKa = 5.96GRR206 pKa = 11.84YY207 pKa = 9.75LSDD210 pKa = 4.41HH211 pKa = 6.32IVGKK215 pKa = 9.7GIRR218 pKa = 11.84GMSLLAWGRR227 pKa = 11.84SKK229 pKa = 9.78TGKK232 pKa = 8.59TMWARR237 pKa = 11.84SLGKK241 pKa = 9.22HH242 pKa = 5.95AYY244 pKa = 9.85FGGLFSLDD252 pKa = 2.85EE253 pKa = 4.24WEE255 pKa = 4.07QDD257 pKa = 3.12EE258 pKa = 4.69SVDD261 pKa = 3.64YY262 pKa = 11.3AVFDD266 pKa = 5.29DD267 pKa = 3.86IQGGFKK273 pKa = 10.47FFPAYY278 pKa = 9.75KK279 pKa = 9.8SWLGQQTEE287 pKa = 4.45FHH289 pKa = 6.2CTDD292 pKa = 3.53RR293 pKa = 11.84YY294 pKa = 10.06RR295 pKa = 11.84KK296 pKa = 9.4KK297 pKa = 10.77KK298 pKa = 8.75HH299 pKa = 4.86VQWGKK304 pKa = 8.63PCIWLMNEE312 pKa = 4.66DD313 pKa = 5.19PYY315 pKa = 11.33QQDD318 pKa = 3.06VDD320 pKa = 4.11IDD322 pKa = 3.65WLEE325 pKa = 4.12ANCLIVNVDD334 pKa = 3.02NSLFF338 pKa = 3.65
Molecular weight: 39.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3M1X5|A0A0E3M1X5_9VIRU Rep OS=Mongoose feces-associated gemycircularvirus b OX=1634486 PE=3 SV=1
MM1 pKa = 6.95THH3 pKa = 6.2SKK5 pKa = 8.6MAYY8 pKa = 7.83RR9 pKa = 11.84SRR11 pKa = 11.84RR12 pKa = 11.84FTGRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GFTPRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SSFRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84SFRR35 pKa = 11.84SRR37 pKa = 11.84PTRR40 pKa = 11.84NYY42 pKa = 9.69RR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84TNVRR50 pKa = 11.84RR51 pKa = 11.84VRR53 pKa = 11.84NLASRR58 pKa = 11.84KK59 pKa = 9.83CKK61 pKa = 10.56DD62 pKa = 3.39SMLSTPIDD70 pKa = 3.5DD71 pKa = 3.98TGTPEE76 pKa = 4.05TPGPVSMNAANVYY89 pKa = 10.06GFIYY93 pKa = 10.36SPSARR98 pKa = 11.84VAGHH102 pKa = 6.96LLDD105 pKa = 3.83SGEE108 pKa = 4.25RR109 pKa = 11.84VNTPSHH115 pKa = 4.48SQRR118 pKa = 11.84RR119 pKa = 11.84KK120 pKa = 7.75SKK122 pKa = 10.46CYY124 pKa = 8.68VRR126 pKa = 11.84GYY128 pKa = 10.86GEE130 pKa = 4.14TVEE133 pKa = 4.56IQTDD137 pKa = 3.97DD138 pKa = 3.42ASPWIWRR145 pKa = 11.84RR146 pKa = 11.84IVFSTIGLAVQFSPGLLYY164 pKa = 11.29NNDD167 pKa = 3.29DD168 pKa = 3.31TRR170 pKa = 11.84GYY172 pKa = 10.07GRR174 pKa = 11.84TMYY177 pKa = 10.34NLRR180 pKa = 11.84SGTPQADD187 pKa = 3.56TLEE190 pKa = 5.0GIVYY194 pKa = 9.99RR195 pKa = 11.84YY196 pKa = 9.79LFQGTQDD203 pKa = 3.39IDD205 pKa = 3.25WANVMTATTAPRR217 pKa = 11.84HH218 pKa = 5.06VKK220 pKa = 10.05VYY222 pKa = 9.86SDD224 pKa = 3.52RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84IIQSPNSDD235 pKa = 2.92GGTIRR240 pKa = 11.84ISKK243 pKa = 10.11HH244 pKa = 4.6YY245 pKa = 10.66DD246 pKa = 3.46GINRR250 pKa = 11.84SMIYY254 pKa = 10.72DD255 pKa = 3.79DD256 pKa = 4.44VEE258 pKa = 5.62AGAGPKK264 pKa = 8.62ATNSIASGTPYY275 pKa = 11.34GNMGDD280 pKa = 4.75LFVLDD285 pKa = 4.95FFSSASDD292 pKa = 4.04DD293 pKa = 3.89EE294 pKa = 4.71TSGATFQPHH303 pKa = 4.72GRR305 pKa = 11.84YY306 pKa = 8.68YY307 pKa = 8.23WHH309 pKa = 7.47EE310 pKa = 4.37GQAHH314 pKa = 6.07
MM1 pKa = 6.95THH3 pKa = 6.2SKK5 pKa = 8.6MAYY8 pKa = 7.83RR9 pKa = 11.84SRR11 pKa = 11.84RR12 pKa = 11.84FTGRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GFTPRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SSFRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84SFRR35 pKa = 11.84SRR37 pKa = 11.84PTRR40 pKa = 11.84NYY42 pKa = 9.69RR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84TNVRR50 pKa = 11.84RR51 pKa = 11.84VRR53 pKa = 11.84NLASRR58 pKa = 11.84KK59 pKa = 9.83CKK61 pKa = 10.56DD62 pKa = 3.39SMLSTPIDD70 pKa = 3.5DD71 pKa = 3.98TGTPEE76 pKa = 4.05TPGPVSMNAANVYY89 pKa = 10.06GFIYY93 pKa = 10.36SPSARR98 pKa = 11.84VAGHH102 pKa = 6.96LLDD105 pKa = 3.83SGEE108 pKa = 4.25RR109 pKa = 11.84VNTPSHH115 pKa = 4.48SQRR118 pKa = 11.84RR119 pKa = 11.84KK120 pKa = 7.75SKK122 pKa = 10.46CYY124 pKa = 8.68VRR126 pKa = 11.84GYY128 pKa = 10.86GEE130 pKa = 4.14TVEE133 pKa = 4.56IQTDD137 pKa = 3.97DD138 pKa = 3.42ASPWIWRR145 pKa = 11.84RR146 pKa = 11.84IVFSTIGLAVQFSPGLLYY164 pKa = 11.29NNDD167 pKa = 3.29DD168 pKa = 3.31TRR170 pKa = 11.84GYY172 pKa = 10.07GRR174 pKa = 11.84TMYY177 pKa = 10.34NLRR180 pKa = 11.84SGTPQADD187 pKa = 3.56TLEE190 pKa = 5.0GIVYY194 pKa = 9.99RR195 pKa = 11.84YY196 pKa = 9.79LFQGTQDD203 pKa = 3.39IDD205 pKa = 3.25WANVMTATTAPRR217 pKa = 11.84HH218 pKa = 5.06VKK220 pKa = 10.05VYY222 pKa = 9.86SDD224 pKa = 3.52RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84IIQSPNSDD235 pKa = 2.92GGTIRR240 pKa = 11.84ISKK243 pKa = 10.11HH244 pKa = 4.6YY245 pKa = 10.66DD246 pKa = 3.46GINRR250 pKa = 11.84SMIYY254 pKa = 10.72DD255 pKa = 3.79DD256 pKa = 4.44VEE258 pKa = 5.62AGAGPKK264 pKa = 8.62ATNSIASGTPYY275 pKa = 11.34GNMGDD280 pKa = 4.75LFVLDD285 pKa = 4.95FFSSASDD292 pKa = 4.04DD293 pKa = 3.89EE294 pKa = 4.71TSGATFQPHH303 pKa = 4.72GRR305 pKa = 11.84YY306 pKa = 8.68YY307 pKa = 8.23WHH309 pKa = 7.47EE310 pKa = 4.37GQAHH314 pKa = 6.07
Molecular weight: 35.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
652 |
314 |
338 |
326.0 |
37.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.675 ± 0.266 | 1.227 ± 0.418 |
7.362 ± 0.703 | 3.681 ± 0.803 |
5.521 ± 1.204 | 8.436 ± 0.116 |
3.528 ± 0.694 | 4.294 ± 0.342 |
3.834 ± 0.911 | 5.061 ± 0.878 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.994 ± 0.392 | 3.988 ± 0.334 |
4.448 ± 0.684 | 3.681 ± 0.577 |
10.583 ± 1.753 | 8.282 ± 1.126 |
5.982 ± 1.628 | 4.755 ± 0.241 |
2.454 ± 0.836 | 5.215 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
