
Avian metapneumovirus type D
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Metapneumovirus; Avian metapneumovirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
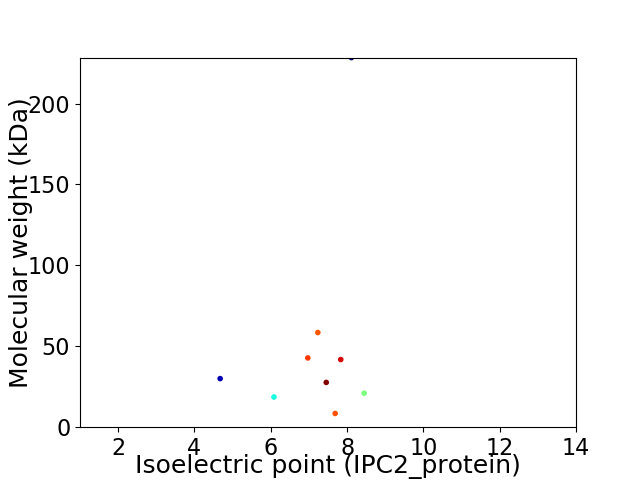
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077SG97|A0A077SG97_9MONO Small hydrophobic protein OS=Avian metapneumovirus type D OX=519376 GN=SH PE=4 SV=1
MM1 pKa = 7.55SFPEE5 pKa = 4.55GKK7 pKa = 9.88DD8 pKa = 2.94ILMMGSEE15 pKa = 4.21AAKK18 pKa = 10.53LADD21 pKa = 4.64AYY23 pKa = 10.85QKK25 pKa = 10.73SLKK28 pKa = 10.65DD29 pKa = 3.26PSRR32 pKa = 11.84NSKK35 pKa = 10.48SISGDD40 pKa = 3.12PVATVSEE47 pKa = 4.43RR48 pKa = 11.84VPTLPLCGPISPKK61 pKa = 10.54GSSTKK66 pKa = 8.65PTRR69 pKa = 11.84EE70 pKa = 3.66ASPAPKK76 pKa = 8.78KK77 pKa = 9.24TGPIYY82 pKa = 10.25PKK84 pKa = 10.66LPTAPPDD91 pKa = 4.01PGQPEE96 pKa = 5.27PITSNGPKK104 pKa = 9.9KK105 pKa = 10.24AQKK108 pKa = 9.76KK109 pKa = 8.26VKK111 pKa = 10.08FDD113 pKa = 3.34AVKK116 pKa = 9.64PGKK119 pKa = 9.76YY120 pKa = 9.73SKK122 pKa = 11.09LEE124 pKa = 3.99EE125 pKa = 4.16EE126 pKa = 4.31ALEE129 pKa = 4.25LLSDD133 pKa = 4.23PDD135 pKa = 3.71EE136 pKa = 4.92EE137 pKa = 4.27NDD139 pKa = 3.8KK140 pKa = 10.95EE141 pKa = 4.56SSILTFEE148 pKa = 4.56EE149 pKa = 4.09KK150 pKa = 10.8DD151 pKa = 3.5NASTSIEE158 pKa = 3.89ARR160 pKa = 11.84LEE162 pKa = 4.34AIEE165 pKa = 4.92EE166 pKa = 4.24KK167 pKa = 10.89LSMILGMLKK176 pKa = 9.1TLSIATAGPTAARR189 pKa = 11.84DD190 pKa = 4.19GIRR193 pKa = 11.84DD194 pKa = 3.36AMVGMRR200 pKa = 11.84EE201 pKa = 3.93EE202 pKa = 5.1LINSIMAEE210 pKa = 3.98AKK212 pKa = 10.55DD213 pKa = 4.7KK214 pKa = 10.4ISEE217 pKa = 4.27MIKK220 pKa = 10.67EE221 pKa = 4.18EE222 pKa = 3.77DD223 pKa = 3.39TQRR226 pKa = 11.84AKK228 pKa = 10.66IGDD231 pKa = 3.48GSVRR235 pKa = 11.84LTEE238 pKa = 4.4KK239 pKa = 10.74AKK241 pKa = 10.41EE242 pKa = 3.96LNKK245 pKa = 10.04VLEE248 pKa = 4.79DD249 pKa = 3.49QSSSGEE255 pKa = 3.99SDD257 pKa = 3.5SEE259 pKa = 4.72SEE261 pKa = 4.5DD262 pKa = 3.81EE263 pKa = 4.39EE264 pKa = 4.6PEE266 pKa = 4.06PDD268 pKa = 3.46EE269 pKa = 4.26TADD272 pKa = 3.54IYY274 pKa = 11.54SLEE277 pKa = 4.07LL278 pKa = 4.26
MM1 pKa = 7.55SFPEE5 pKa = 4.55GKK7 pKa = 9.88DD8 pKa = 2.94ILMMGSEE15 pKa = 4.21AAKK18 pKa = 10.53LADD21 pKa = 4.64AYY23 pKa = 10.85QKK25 pKa = 10.73SLKK28 pKa = 10.65DD29 pKa = 3.26PSRR32 pKa = 11.84NSKK35 pKa = 10.48SISGDD40 pKa = 3.12PVATVSEE47 pKa = 4.43RR48 pKa = 11.84VPTLPLCGPISPKK61 pKa = 10.54GSSTKK66 pKa = 8.65PTRR69 pKa = 11.84EE70 pKa = 3.66ASPAPKK76 pKa = 8.78KK77 pKa = 9.24TGPIYY82 pKa = 10.25PKK84 pKa = 10.66LPTAPPDD91 pKa = 4.01PGQPEE96 pKa = 5.27PITSNGPKK104 pKa = 9.9KK105 pKa = 10.24AQKK108 pKa = 9.76KK109 pKa = 8.26VKK111 pKa = 10.08FDD113 pKa = 3.34AVKK116 pKa = 9.64PGKK119 pKa = 9.76YY120 pKa = 9.73SKK122 pKa = 11.09LEE124 pKa = 3.99EE125 pKa = 4.16EE126 pKa = 4.31ALEE129 pKa = 4.25LLSDD133 pKa = 4.23PDD135 pKa = 3.71EE136 pKa = 4.92EE137 pKa = 4.27NDD139 pKa = 3.8KK140 pKa = 10.95EE141 pKa = 4.56SSILTFEE148 pKa = 4.56EE149 pKa = 4.09KK150 pKa = 10.8DD151 pKa = 3.5NASTSIEE158 pKa = 3.89ARR160 pKa = 11.84LEE162 pKa = 4.34AIEE165 pKa = 4.92EE166 pKa = 4.24KK167 pKa = 10.89LSMILGMLKK176 pKa = 9.1TLSIATAGPTAARR189 pKa = 11.84DD190 pKa = 4.19GIRR193 pKa = 11.84DD194 pKa = 3.36AMVGMRR200 pKa = 11.84EE201 pKa = 3.93EE202 pKa = 5.1LINSIMAEE210 pKa = 3.98AKK212 pKa = 10.55DD213 pKa = 4.7KK214 pKa = 10.4ISEE217 pKa = 4.27MIKK220 pKa = 10.67EE221 pKa = 4.18EE222 pKa = 3.77DD223 pKa = 3.39TQRR226 pKa = 11.84AKK228 pKa = 10.66IGDD231 pKa = 3.48GSVRR235 pKa = 11.84LTEE238 pKa = 4.4KK239 pKa = 10.74AKK241 pKa = 10.41EE242 pKa = 3.96LNKK245 pKa = 10.04VLEE248 pKa = 4.79DD249 pKa = 3.49QSSSGEE255 pKa = 3.99SDD257 pKa = 3.5SEE259 pKa = 4.72SEE261 pKa = 4.5DD262 pKa = 3.81EE263 pKa = 4.39EE264 pKa = 4.6PEE266 pKa = 4.06PDD268 pKa = 3.46EE269 pKa = 4.26TADD272 pKa = 3.54IYY274 pKa = 11.54SLEE277 pKa = 4.07LL278 pKa = 4.26
Molecular weight: 30.04 kDa
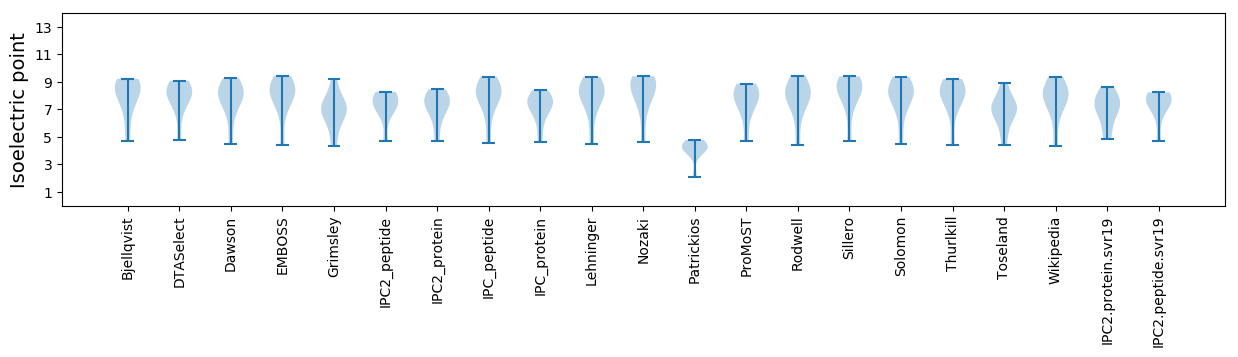
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077SJ11|A0A077SJ11_9MONO Matrix protein 2-2 OS=Avian metapneumovirus type D OX=519376 GN=M2.2 PE=4 SV=1
MM1 pKa = 7.68SKK3 pKa = 10.5RR4 pKa = 11.84NPCRR8 pKa = 11.84YY9 pKa = 9.76EE10 pKa = 3.46IRR12 pKa = 11.84GKK14 pKa = 9.97CNRR17 pKa = 11.84GSSCSFNHH25 pKa = 6.62NYY27 pKa = 9.05WSWPDD32 pKa = 3.23HH33 pKa = 5.68VLLVRR38 pKa = 11.84INCMLNQLQRR48 pKa = 11.84NTDD51 pKa = 3.42RR52 pKa = 11.84TDD54 pKa = 3.29GLSLISGAGRR64 pKa = 11.84EE65 pKa = 4.33DD66 pKa = 3.38RR67 pKa = 11.84TQDD70 pKa = 3.55FVLGSANVVQGYY82 pKa = 10.09IDD84 pKa = 4.0GNATITKK91 pKa = 9.99SAACYY96 pKa = 10.26SLYY99 pKa = 10.99NIIKK103 pKa = 9.6QLQEE107 pKa = 3.8NDD109 pKa = 3.15VKK111 pKa = 11.08ASRR114 pKa = 11.84DD115 pKa = 3.5TMLDD119 pKa = 3.33DD120 pKa = 4.53HH121 pKa = 6.76KK122 pKa = 10.69HH123 pKa = 4.69VALHH127 pKa = 6.1NLVLSYY133 pKa = 10.97IDD135 pKa = 3.46MSKK138 pKa = 10.78NPASLINSLKK148 pKa = 10.5RR149 pKa = 11.84LPKK152 pKa = 10.42EE153 pKa = 3.91KK154 pKa = 10.38LKK156 pKa = 11.01KK157 pKa = 8.17LAKK160 pKa = 10.54VIVQLSAGPEE170 pKa = 3.82VEE172 pKa = 4.32NASGHH177 pKa = 5.82PMQKK181 pKa = 10.09GDD183 pKa = 4.43SGHH186 pKa = 5.42QVV188 pKa = 2.65
MM1 pKa = 7.68SKK3 pKa = 10.5RR4 pKa = 11.84NPCRR8 pKa = 11.84YY9 pKa = 9.76EE10 pKa = 3.46IRR12 pKa = 11.84GKK14 pKa = 9.97CNRR17 pKa = 11.84GSSCSFNHH25 pKa = 6.62NYY27 pKa = 9.05WSWPDD32 pKa = 3.23HH33 pKa = 5.68VLLVRR38 pKa = 11.84INCMLNQLQRR48 pKa = 11.84NTDD51 pKa = 3.42RR52 pKa = 11.84TDD54 pKa = 3.29GLSLISGAGRR64 pKa = 11.84EE65 pKa = 4.33DD66 pKa = 3.38RR67 pKa = 11.84TQDD70 pKa = 3.55FVLGSANVVQGYY82 pKa = 10.09IDD84 pKa = 4.0GNATITKK91 pKa = 9.99SAACYY96 pKa = 10.26SLYY99 pKa = 10.99NIIKK103 pKa = 9.6QLQEE107 pKa = 3.8NDD109 pKa = 3.15VKK111 pKa = 11.08ASRR114 pKa = 11.84DD115 pKa = 3.5TMLDD119 pKa = 3.33DD120 pKa = 4.53HH121 pKa = 6.76KK122 pKa = 10.69HH123 pKa = 4.69VALHH127 pKa = 6.1NLVLSYY133 pKa = 10.97IDD135 pKa = 3.46MSKK138 pKa = 10.78NPASLINSLKK148 pKa = 10.5RR149 pKa = 11.84LPKK152 pKa = 10.42EE153 pKa = 3.91KK154 pKa = 10.38LKK156 pKa = 11.01KK157 pKa = 8.17LAKK160 pKa = 10.54VIVQLSAGPEE170 pKa = 3.82VEE172 pKa = 4.32NASGHH177 pKa = 5.82PMQKK181 pKa = 10.09GDD183 pKa = 4.43SGHH186 pKa = 5.42QVV188 pKa = 2.65
Molecular weight: 21.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4292 |
73 |
2006 |
476.9 |
53.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.757 ± 0.943 | 2.586 ± 0.601 |
4.497 ± 0.313 | 5.988 ± 0.624 |
2.912 ± 0.604 | 5.685 ± 0.377 |
1.678 ± 0.381 | 6.431 ± 0.36 |
6.267 ± 0.489 | 9.646 ± 0.9 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.889 ± 0.306 | 4.939 ± 0.695 |
4.497 ± 0.891 | 3.518 ± 0.376 |
5.336 ± 0.441 | 8.201 ± 0.469 |
7.106 ± 0.996 | 6.99 ± 0.647 |
0.955 ± 0.309 | 3.122 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
