
Bdellovibrio sp. SKB1291214
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Oligoflexia; Bdellovibrionales; Bdellovibrionaceae; Bdellovibrio; unclassified Bdellovibrio
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
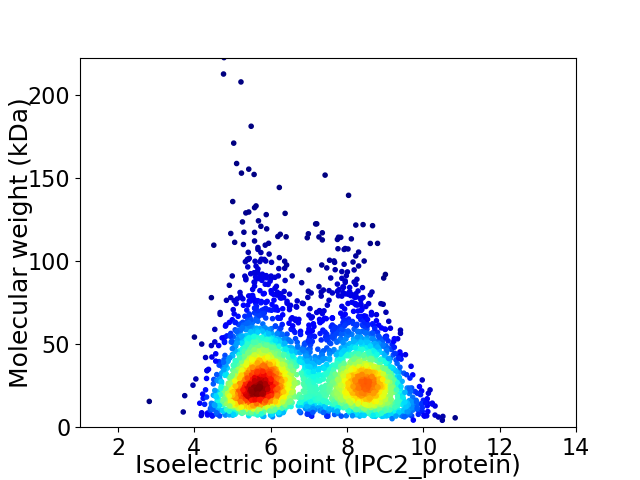
Virtual 2D-PAGE plot for 3588 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A254QHZ5|A0A254QHZ5_9PROT DUF4105 domain-containing protein OS=Bdellovibrio sp. SKB1291214 OX=1732569 GN=B9G69_09260 PE=4 SV=1
MM1 pKa = 6.78VHH3 pKa = 6.6SKK5 pKa = 8.57FTAVLAGLGFLATLSACSAQSDD27 pKa = 3.69KK28 pKa = 11.52DD29 pKa = 3.66KK30 pKa = 11.48LMEE33 pKa = 4.31AQFCLDD39 pKa = 3.42EE40 pKa = 4.47STAATAQSCVSSIAYY55 pKa = 7.35MQTSQSYY62 pKa = 10.84ALQCAAGFIEE72 pKa = 5.18SGVTSPANLSQALDD86 pKa = 3.98SISNNSNSSTALLSALNMGSVSLANNTAEE115 pKa = 4.16YY116 pKa = 9.48CAKK119 pKa = 10.07SGQVGFSLLGAMAKK133 pKa = 9.93SATSIASAASSLGLGSCSTDD153 pKa = 3.48LSQCDD158 pKa = 3.29TSQIEE163 pKa = 4.14DD164 pKa = 4.56TITEE168 pKa = 3.91IQKK171 pKa = 9.2TINSDD176 pKa = 3.35PSATMSLADD185 pKa = 3.23ATAAVEE191 pKa = 4.98AIASSIQTVYY201 pKa = 8.05TTTCGTTGANQDD213 pKa = 2.78ICDD216 pKa = 4.81PIKK219 pKa = 9.98TAIADD224 pKa = 3.74AGVDD228 pKa = 3.21ITDD231 pKa = 4.1PNLDD235 pKa = 3.47LVALGKK241 pKa = 10.27EE242 pKa = 4.97LLAQWKK248 pKa = 8.92PP249 pKa = 3.3
MM1 pKa = 6.78VHH3 pKa = 6.6SKK5 pKa = 8.57FTAVLAGLGFLATLSACSAQSDD27 pKa = 3.69KK28 pKa = 11.52DD29 pKa = 3.66KK30 pKa = 11.48LMEE33 pKa = 4.31AQFCLDD39 pKa = 3.42EE40 pKa = 4.47STAATAQSCVSSIAYY55 pKa = 7.35MQTSQSYY62 pKa = 10.84ALQCAAGFIEE72 pKa = 5.18SGVTSPANLSQALDD86 pKa = 3.98SISNNSNSSTALLSALNMGSVSLANNTAEE115 pKa = 4.16YY116 pKa = 9.48CAKK119 pKa = 10.07SGQVGFSLLGAMAKK133 pKa = 9.93SATSIASAASSLGLGSCSTDD153 pKa = 3.48LSQCDD158 pKa = 3.29TSQIEE163 pKa = 4.14DD164 pKa = 4.56TITEE168 pKa = 3.91IQKK171 pKa = 9.2TINSDD176 pKa = 3.35PSATMSLADD185 pKa = 3.23ATAAVEE191 pKa = 4.98AIASSIQTVYY201 pKa = 8.05TTTCGTTGANQDD213 pKa = 2.78ICDD216 pKa = 4.81PIKK219 pKa = 9.98TAIADD224 pKa = 3.74AGVDD228 pKa = 3.21ITDD231 pKa = 4.1PNLDD235 pKa = 3.47LVALGKK241 pKa = 10.27EE242 pKa = 4.97LLAQWKK248 pKa = 8.92PP249 pKa = 3.3
Molecular weight: 25.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A254QM87|A0A254QM87_9PROT Ribonuclease H OS=Bdellovibrio sp. SKB1291214 OX=1732569 GN=rnhA PE=3 SV=1
MM1 pKa = 7.68NIQEE5 pKa = 4.3NPQRR9 pKa = 11.84APRR12 pKa = 11.84KK13 pKa = 9.43RR14 pKa = 11.84EE15 pKa = 3.7APVPLRR21 pKa = 11.84GPSQDD26 pKa = 2.85LFKK29 pKa = 10.73RR30 pKa = 11.84SKK32 pKa = 10.47QEE34 pKa = 3.54LSFSVGTGFQGGPSRR49 pKa = 11.84KK50 pKa = 8.98RR51 pKa = 11.84KK52 pKa = 9.59GYY54 pKa = 10.5RR55 pKa = 11.84LALWSLMASFIDD67 pKa = 4.39ALILLAISCAFLVLFSLLMKK87 pKa = 10.06TPANAVIKK95 pKa = 10.41IFFRR99 pKa = 11.84SEE101 pKa = 4.0SNIMLLAQLYY111 pKa = 9.77AVSAWVYY118 pKa = 10.21LIATRR123 pKa = 11.84VLTGSSIGEE132 pKa = 3.94WACNIRR138 pKa = 11.84LGQPHH143 pKa = 6.41EE144 pKa = 4.5RR145 pKa = 11.84MEE147 pKa = 4.02TRR149 pKa = 11.84YY150 pKa = 9.18VWRR153 pKa = 11.84VLKK156 pKa = 10.66RR157 pKa = 11.84STLIVATGVVVLPALSLILGGDD179 pKa = 3.75LTGKK183 pKa = 10.31LCGLRR188 pKa = 11.84LFSLKK193 pKa = 10.66
MM1 pKa = 7.68NIQEE5 pKa = 4.3NPQRR9 pKa = 11.84APRR12 pKa = 11.84KK13 pKa = 9.43RR14 pKa = 11.84EE15 pKa = 3.7APVPLRR21 pKa = 11.84GPSQDD26 pKa = 2.85LFKK29 pKa = 10.73RR30 pKa = 11.84SKK32 pKa = 10.47QEE34 pKa = 3.54LSFSVGTGFQGGPSRR49 pKa = 11.84KK50 pKa = 8.98RR51 pKa = 11.84KK52 pKa = 9.59GYY54 pKa = 10.5RR55 pKa = 11.84LALWSLMASFIDD67 pKa = 4.39ALILLAISCAFLVLFSLLMKK87 pKa = 10.06TPANAVIKK95 pKa = 10.41IFFRR99 pKa = 11.84SEE101 pKa = 4.0SNIMLLAQLYY111 pKa = 9.77AVSAWVYY118 pKa = 10.21LIATRR123 pKa = 11.84VLTGSSIGEE132 pKa = 3.94WACNIRR138 pKa = 11.84LGQPHH143 pKa = 6.41EE144 pKa = 4.5RR145 pKa = 11.84MEE147 pKa = 4.02TRR149 pKa = 11.84YY150 pKa = 9.18VWRR153 pKa = 11.84VLKK156 pKa = 10.66RR157 pKa = 11.84STLIVATGVVVLPALSLILGGDD179 pKa = 3.75LTGKK183 pKa = 10.31LCGLRR188 pKa = 11.84LFSLKK193 pKa = 10.66
Molecular weight: 21.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1113114 |
37 |
2152 |
310.2 |
34.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.31 ± 0.039 | 0.918 ± 0.014 |
5.168 ± 0.027 | 6.234 ± 0.048 |
4.573 ± 0.034 | 6.938 ± 0.045 |
1.911 ± 0.02 | 6.081 ± 0.033 |
6.948 ± 0.049 | 9.463 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.801 ± 0.019 | 4.343 ± 0.026 |
4.045 ± 0.026 | 3.954 ± 0.026 |
4.529 ± 0.031 | 6.972 ± 0.044 |
5.581 ± 0.053 | 6.959 ± 0.035 |
1.181 ± 0.016 | 3.091 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
