
Pandoravirus dulcis
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA viruses; Pandoravirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
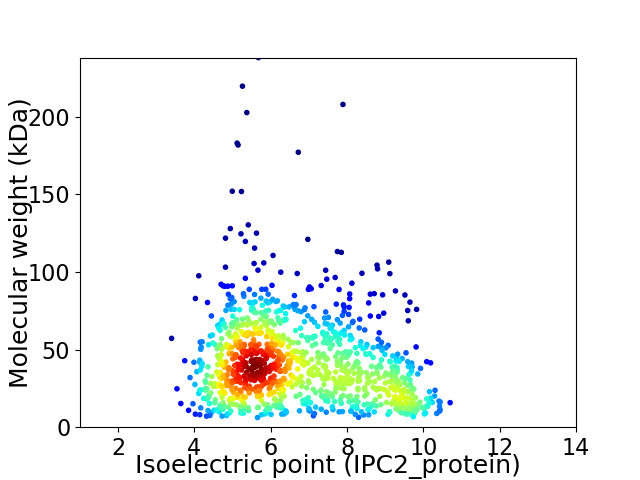
Virtual 2D-PAGE plot for 1070 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4VTA8|S4VTA8_9VIRU Fascin-like protein OS=Pandoravirus dulcis OX=1349409 GN=pdul_cds_543 PE=4 SV=1
MM1 pKa = 7.02ATCSGSPQDD10 pKa = 5.88DD11 pKa = 3.9EE12 pKa = 4.94CCPITLVPFRR22 pKa = 11.84TITAARR28 pKa = 11.84TTFLPCCHH36 pKa = 6.87RR37 pKa = 11.84FDD39 pKa = 4.22SDD41 pKa = 4.8AIRR44 pKa = 11.84HH45 pKa = 5.69YY46 pKa = 10.81IDD48 pKa = 3.23NCLARR53 pKa = 11.84RR54 pKa = 11.84APRR57 pKa = 11.84GAAPPVPCPVCRR69 pKa = 11.84CRR71 pKa = 11.84VPPDD75 pKa = 3.46YY76 pKa = 10.66LADD79 pKa = 3.51MGVPVAWPPSAPVVAPVAAPAVPAAPAVPAAPAASAAIAVAAPATTARR127 pKa = 11.84RR128 pKa = 11.84SSRR131 pKa = 11.84PSRR134 pKa = 11.84PPAPVVIVISDD145 pKa = 4.03DD146 pKa = 4.37DD147 pKa = 4.73DD148 pKa = 6.09DD149 pKa = 5.27SEE151 pKa = 4.5EE152 pKa = 4.07VHH154 pKa = 6.51EE155 pKa = 5.83RR156 pKa = 11.84TAQWINSDD164 pKa = 4.8DD165 pKa = 5.32DD166 pKa = 5.72DD167 pKa = 7.53DD168 pKa = 7.46GDD170 pKa = 6.17DD171 pKa = 6.01DD172 pKa = 7.01DD173 pKa = 7.52DD174 pKa = 6.31DD175 pKa = 6.45DD176 pKa = 4.38STGSLDD182 pKa = 3.92EE183 pKa = 5.41FIVDD187 pKa = 5.3DD188 pKa = 6.21DD189 pKa = 4.2EE190 pKa = 4.64TDD192 pKa = 3.39PDD194 pKa = 4.2AEE196 pKa = 4.29YY197 pKa = 10.38TPAPVVAAPDD207 pKa = 4.41DD208 pKa = 5.4DD209 pKa = 6.0DD210 pKa = 7.58DD211 pKa = 7.08SDD213 pKa = 6.5LDD215 pKa = 5.58DD216 pKa = 6.79DD217 pKa = 6.31DD218 pKa = 7.05DD219 pKa = 6.44DD220 pKa = 4.89STDD223 pKa = 3.76SDD225 pKa = 4.8CSCLTDD231 pKa = 4.66SEE233 pKa = 5.35DD234 pKa = 3.16II235 pKa = 4.24
MM1 pKa = 7.02ATCSGSPQDD10 pKa = 5.88DD11 pKa = 3.9EE12 pKa = 4.94CCPITLVPFRR22 pKa = 11.84TITAARR28 pKa = 11.84TTFLPCCHH36 pKa = 6.87RR37 pKa = 11.84FDD39 pKa = 4.22SDD41 pKa = 4.8AIRR44 pKa = 11.84HH45 pKa = 5.69YY46 pKa = 10.81IDD48 pKa = 3.23NCLARR53 pKa = 11.84RR54 pKa = 11.84APRR57 pKa = 11.84GAAPPVPCPVCRR69 pKa = 11.84CRR71 pKa = 11.84VPPDD75 pKa = 3.46YY76 pKa = 10.66LADD79 pKa = 3.51MGVPVAWPPSAPVVAPVAAPAVPAAPAVPAAPAASAAIAVAAPATTARR127 pKa = 11.84RR128 pKa = 11.84SSRR131 pKa = 11.84PSRR134 pKa = 11.84PPAPVVIVISDD145 pKa = 4.03DD146 pKa = 4.37DD147 pKa = 4.73DD148 pKa = 6.09DD149 pKa = 5.27SEE151 pKa = 4.5EE152 pKa = 4.07VHH154 pKa = 6.51EE155 pKa = 5.83RR156 pKa = 11.84TAQWINSDD164 pKa = 4.8DD165 pKa = 5.32DD166 pKa = 5.72DD167 pKa = 7.53DD168 pKa = 7.46GDD170 pKa = 6.17DD171 pKa = 6.01DD172 pKa = 7.01DD173 pKa = 7.52DD174 pKa = 6.31DD175 pKa = 6.45DD176 pKa = 4.38STGSLDD182 pKa = 3.92EE183 pKa = 5.41FIVDD187 pKa = 5.3DD188 pKa = 6.21DD189 pKa = 4.2EE190 pKa = 4.64TDD192 pKa = 3.39PDD194 pKa = 4.2AEE196 pKa = 4.29YY197 pKa = 10.38TPAPVVAAPDD207 pKa = 4.41DD208 pKa = 5.4DD209 pKa = 6.0DD210 pKa = 7.58DD211 pKa = 7.08SDD213 pKa = 6.5LDD215 pKa = 5.58DD216 pKa = 6.79DD217 pKa = 6.31DD218 pKa = 7.05DD219 pKa = 6.44DD220 pKa = 4.89STDD223 pKa = 3.76SDD225 pKa = 4.8CSCLTDD231 pKa = 4.66SEE233 pKa = 5.35DD234 pKa = 3.16II235 pKa = 4.24
Molecular weight: 24.75 kDa
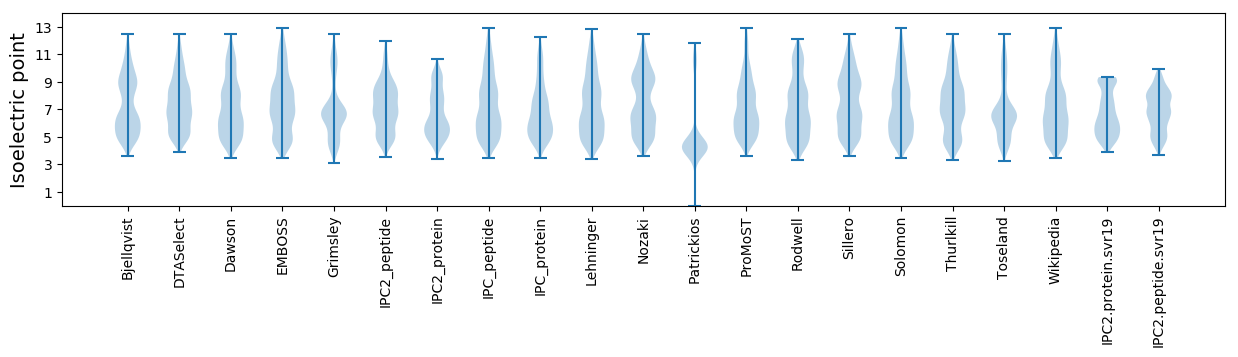
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4VSA2|S4VSA2_9VIRU Uncharacterized protein OS=Pandoravirus dulcis OX=1349409 GN=pdul_cds_935 PE=4 SV=1
MM1 pKa = 7.97GDD3 pKa = 3.41RR4 pKa = 11.84PDD6 pKa = 4.5RR7 pKa = 11.84GSKK10 pKa = 8.38CTRR13 pKa = 11.84APAFVVGSCVCTEE26 pKa = 4.14RR27 pKa = 11.84EE28 pKa = 4.26VRR30 pKa = 11.84WSFFLLVVILTLVTISLVLFVSAVRR55 pKa = 11.84YY56 pKa = 7.57VKK58 pKa = 10.34SWVSEE63 pKa = 4.08KK64 pKa = 10.33QRR66 pKa = 11.84LRR68 pKa = 11.84ADD70 pKa = 3.43VKK72 pKa = 10.86ALSRR76 pKa = 11.84QASGVMRR83 pKa = 11.84DD84 pKa = 3.48AQQRR88 pKa = 11.84AAAAASAPSHH98 pKa = 6.56VPDD101 pKa = 4.13ATATAPPARR110 pKa = 11.84APPSAAEE117 pKa = 3.8SAAASKK123 pKa = 10.96ARR125 pKa = 11.84TAAVAAADD133 pKa = 3.98PLVHH137 pKa = 6.59EE138 pKa = 4.82MLDD141 pKa = 3.61AIEE144 pKa = 4.02AVRR147 pKa = 11.84LGSCGRR153 pKa = 11.84GFAFAVDD160 pKa = 3.53SRR162 pKa = 11.84GGVWAHH168 pKa = 5.67GKK170 pKa = 7.39TRR172 pKa = 11.84HH173 pKa = 5.65LARR176 pKa = 11.84GATGRR181 pKa = 11.84VPGFHH186 pKa = 6.56RR187 pKa = 11.84VADD190 pKa = 4.1ADD192 pKa = 3.8HH193 pKa = 6.93TGGSTSGAYY202 pKa = 10.22DD203 pKa = 3.89CDD205 pKa = 4.41DD206 pKa = 3.89DD207 pKa = 5.93HH208 pKa = 7.06EE209 pKa = 7.06ADD211 pKa = 3.26ASVYY215 pKa = 10.54GGAPVARR222 pKa = 11.84GRR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84GGAPARR233 pKa = 11.84APSPLADD240 pKa = 3.4MLGAARR246 pKa = 11.84RR247 pKa = 11.84GGGYY251 pKa = 7.49VHH253 pKa = 6.79FRR255 pKa = 11.84WKK257 pKa = 10.73RR258 pKa = 11.84EE259 pKa = 4.0VLMMAYY265 pKa = 8.59VHH267 pKa = 6.15PVKK270 pKa = 10.37GTDD273 pKa = 3.4LVLGGAVPVPRR284 pKa = 11.84KK285 pKa = 9.16QKK287 pKa = 10.36AWEE290 pKa = 3.96EE291 pKa = 4.05ANAARR296 pKa = 11.84PAAPARR302 pKa = 11.84RR303 pKa = 3.79
MM1 pKa = 7.97GDD3 pKa = 3.41RR4 pKa = 11.84PDD6 pKa = 4.5RR7 pKa = 11.84GSKK10 pKa = 8.38CTRR13 pKa = 11.84APAFVVGSCVCTEE26 pKa = 4.14RR27 pKa = 11.84EE28 pKa = 4.26VRR30 pKa = 11.84WSFFLLVVILTLVTISLVLFVSAVRR55 pKa = 11.84YY56 pKa = 7.57VKK58 pKa = 10.34SWVSEE63 pKa = 4.08KK64 pKa = 10.33QRR66 pKa = 11.84LRR68 pKa = 11.84ADD70 pKa = 3.43VKK72 pKa = 10.86ALSRR76 pKa = 11.84QASGVMRR83 pKa = 11.84DD84 pKa = 3.48AQQRR88 pKa = 11.84AAAAASAPSHH98 pKa = 6.56VPDD101 pKa = 4.13ATATAPPARR110 pKa = 11.84APPSAAEE117 pKa = 3.8SAAASKK123 pKa = 10.96ARR125 pKa = 11.84TAAVAAADD133 pKa = 3.98PLVHH137 pKa = 6.59EE138 pKa = 4.82MLDD141 pKa = 3.61AIEE144 pKa = 4.02AVRR147 pKa = 11.84LGSCGRR153 pKa = 11.84GFAFAVDD160 pKa = 3.53SRR162 pKa = 11.84GGVWAHH168 pKa = 5.67GKK170 pKa = 7.39TRR172 pKa = 11.84HH173 pKa = 5.65LARR176 pKa = 11.84GATGRR181 pKa = 11.84VPGFHH186 pKa = 6.56RR187 pKa = 11.84VADD190 pKa = 4.1ADD192 pKa = 3.8HH193 pKa = 6.93TGGSTSGAYY202 pKa = 10.22DD203 pKa = 3.89CDD205 pKa = 4.41DD206 pKa = 3.89DD207 pKa = 5.93HH208 pKa = 7.06EE209 pKa = 7.06ADD211 pKa = 3.26ASVYY215 pKa = 10.54GGAPVARR222 pKa = 11.84GRR224 pKa = 11.84RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84GGAPARR233 pKa = 11.84APSPLADD240 pKa = 3.4MLGAARR246 pKa = 11.84RR247 pKa = 11.84GGGYY251 pKa = 7.49VHH253 pKa = 6.79FRR255 pKa = 11.84WKK257 pKa = 10.73RR258 pKa = 11.84EE259 pKa = 4.0VLMMAYY265 pKa = 8.59VHH267 pKa = 6.15PVKK270 pKa = 10.37GTDD273 pKa = 3.4LVLGGAVPVPRR284 pKa = 11.84KK285 pKa = 9.16QKK287 pKa = 10.36AWEE290 pKa = 3.96EE291 pKa = 4.05ANAARR296 pKa = 11.84PAAPARR302 pKa = 11.84RR303 pKa = 3.79
Molecular weight: 31.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
424486 |
60 |
2245 |
396.7 |
42.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.358 ± 0.097 | 2.433 ± 0.044 |
7.623 ± 0.079 | 3.921 ± 0.049 |
2.213 ± 0.033 | 8.227 ± 0.071 |
2.788 ± 0.046 | 2.809 ± 0.031 |
1.879 ± 0.048 | 7.964 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.911 ± 0.026 | 1.84 ± 0.031 |
6.873 ± 0.081 | 2.469 ± 0.039 |
9.165 ± 0.069 | 5.629 ± 0.062 |
5.51 ± 0.056 | 7.519 ± 0.064 |
1.863 ± 0.036 | 2.005 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
