
Lactobacillus gastricus PS3
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Limosilactobacillus; Limosilactobacillus gastricus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
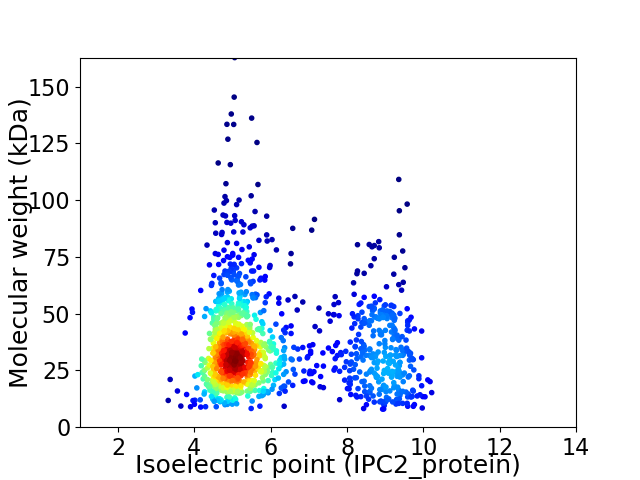
Virtual 2D-PAGE plot for 1269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H4GHT9|H4GHT9_9LACO DNA-binding protein HU OS=Lactobacillus gastricus PS3 OX=1144300 GN=PS3_15348 PE=3 SV=1
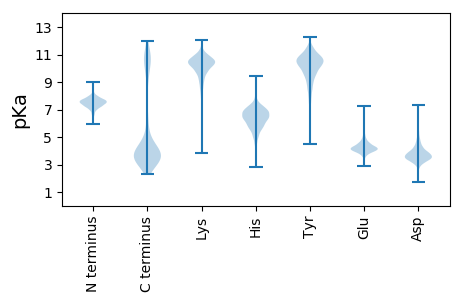
MM1 pKa = 7.47SYY3 pKa = 10.38SEE5 pKa = 5.23KK6 pKa = 10.45MLDD9 pKa = 3.47QLDD12 pKa = 3.76RR13 pKa = 11.84GQLAEE18 pKa = 4.31AKK20 pKa = 10.37KK21 pKa = 10.42SFAWALRR28 pKa = 11.84KK29 pKa = 10.11DD30 pKa = 3.75DD31 pKa = 6.3DD32 pKa = 3.78EE33 pKa = 4.66TLFNLAEE40 pKa = 4.03EE41 pKa = 4.9LYY43 pKa = 11.26GLGFLNQAQRR53 pKa = 11.84VYY55 pKa = 11.27LKK57 pKa = 10.86LLEE60 pKa = 4.81KK61 pKa = 11.14YY62 pKa = 9.81PDD64 pKa = 3.52EE65 pKa = 5.03DD66 pKa = 3.69VLRR69 pKa = 11.84TALAEE74 pKa = 3.82IAIDD78 pKa = 3.61NDD80 pKa = 3.94HH81 pKa = 6.91NDD83 pKa = 3.29EE84 pKa = 4.39ALTYY88 pKa = 9.97LNQIAADD95 pKa = 3.49SDD97 pKa = 4.23AYY99 pKa = 11.29VPALLVAADD108 pKa = 4.79LYY110 pKa = 9.64QTEE113 pKa = 4.31EE114 pKa = 3.95QFEE117 pKa = 4.44VTEE120 pKa = 4.39SKK122 pKa = 10.85LLEE125 pKa = 4.71AYY127 pKa = 10.13QIAPDD132 pKa = 3.73EE133 pKa = 4.34PAVLFALGEE142 pKa = 4.19FYY144 pKa = 11.42NLIGKK149 pKa = 8.46PEE151 pKa = 3.87QAIQYY156 pKa = 8.67YY157 pKa = 9.55FALIQAGYY165 pKa = 10.92LEE167 pKa = 4.28FAKK170 pKa = 10.67VDD172 pKa = 3.01IAGRR176 pKa = 11.84LGTAYY181 pKa = 10.06AQSGHH186 pKa = 6.36FDD188 pKa = 3.05QALGYY193 pKa = 10.04LEE195 pKa = 5.13QVDD198 pKa = 4.18PQYY201 pKa = 11.06QSSDD205 pKa = 2.61IRR207 pKa = 11.84FQTGFTQLQLGKK219 pKa = 10.4LKK221 pKa = 10.6AAEE224 pKa = 4.1QSLQSLIDD232 pKa = 4.55DD233 pKa = 4.6DD234 pKa = 4.24SQYY237 pKa = 11.87ASAYY241 pKa = 7.0PALATVYY248 pKa = 9.53EE249 pKa = 4.12QEE251 pKa = 4.19NDD253 pKa = 3.31YY254 pKa = 10.96QQALKK259 pKa = 9.42TVQEE263 pKa = 4.35GLSVDD268 pKa = 3.66QYY270 pKa = 12.02NEE272 pKa = 3.55NLYY275 pKa = 10.78AQAADD280 pKa = 3.96YY281 pKa = 10.86ASHH284 pKa = 7.13LGDD287 pKa = 5.36DD288 pKa = 4.31PLMDD292 pKa = 3.41QYY294 pKa = 11.39LQRR297 pKa = 11.84AHH299 pKa = 7.24EE300 pKa = 4.45IDD302 pKa = 4.37PDD304 pKa = 3.66NLTITIQYY312 pKa = 11.13SNFLLSQQRR321 pKa = 11.84DD322 pKa = 3.44QDD324 pKa = 4.0NLNLLASVEE333 pKa = 4.1DD334 pKa = 4.53DD335 pKa = 3.54EE336 pKa = 6.03VVDD339 pKa = 5.68PQVEE343 pKa = 4.16WNRR346 pKa = 11.84AQSYY350 pKa = 7.82WRR352 pKa = 11.84LEE354 pKa = 3.98QFDD357 pKa = 3.63QAGQAFSAALPAFEE371 pKa = 5.86DD372 pKa = 3.76NPDD375 pKa = 3.95FLHH378 pKa = 6.77QLIDD382 pKa = 3.5YY383 pKa = 7.54YY384 pKa = 10.92QAVGEE389 pKa = 4.2RR390 pKa = 11.84DD391 pKa = 3.44LMFDD395 pKa = 2.93QLQRR399 pKa = 11.84YY400 pKa = 8.44VEE402 pKa = 4.19LVPTDD407 pKa = 3.23TEE409 pKa = 4.03MAEE412 pKa = 3.98RR413 pKa = 11.84LAEE416 pKa = 4.88FEE418 pKa = 4.26DD419 pKa = 3.83QMYY422 pKa = 10.91
MM1 pKa = 7.47SYY3 pKa = 10.38SEE5 pKa = 5.23KK6 pKa = 10.45MLDD9 pKa = 3.47QLDD12 pKa = 3.76RR13 pKa = 11.84GQLAEE18 pKa = 4.31AKK20 pKa = 10.37KK21 pKa = 10.42SFAWALRR28 pKa = 11.84KK29 pKa = 10.11DD30 pKa = 3.75DD31 pKa = 6.3DD32 pKa = 3.78EE33 pKa = 4.66TLFNLAEE40 pKa = 4.03EE41 pKa = 4.9LYY43 pKa = 11.26GLGFLNQAQRR53 pKa = 11.84VYY55 pKa = 11.27LKK57 pKa = 10.86LLEE60 pKa = 4.81KK61 pKa = 11.14YY62 pKa = 9.81PDD64 pKa = 3.52EE65 pKa = 5.03DD66 pKa = 3.69VLRR69 pKa = 11.84TALAEE74 pKa = 3.82IAIDD78 pKa = 3.61NDD80 pKa = 3.94HH81 pKa = 6.91NDD83 pKa = 3.29EE84 pKa = 4.39ALTYY88 pKa = 9.97LNQIAADD95 pKa = 3.49SDD97 pKa = 4.23AYY99 pKa = 11.29VPALLVAADD108 pKa = 4.79LYY110 pKa = 9.64QTEE113 pKa = 4.31EE114 pKa = 3.95QFEE117 pKa = 4.44VTEE120 pKa = 4.39SKK122 pKa = 10.85LLEE125 pKa = 4.71AYY127 pKa = 10.13QIAPDD132 pKa = 3.73EE133 pKa = 4.34PAVLFALGEE142 pKa = 4.19FYY144 pKa = 11.42NLIGKK149 pKa = 8.46PEE151 pKa = 3.87QAIQYY156 pKa = 8.67YY157 pKa = 9.55FALIQAGYY165 pKa = 10.92LEE167 pKa = 4.28FAKK170 pKa = 10.67VDD172 pKa = 3.01IAGRR176 pKa = 11.84LGTAYY181 pKa = 10.06AQSGHH186 pKa = 6.36FDD188 pKa = 3.05QALGYY193 pKa = 10.04LEE195 pKa = 5.13QVDD198 pKa = 4.18PQYY201 pKa = 11.06QSSDD205 pKa = 2.61IRR207 pKa = 11.84FQTGFTQLQLGKK219 pKa = 10.4LKK221 pKa = 10.6AAEE224 pKa = 4.1QSLQSLIDD232 pKa = 4.55DD233 pKa = 4.6DD234 pKa = 4.24SQYY237 pKa = 11.87ASAYY241 pKa = 7.0PALATVYY248 pKa = 9.53EE249 pKa = 4.12QEE251 pKa = 4.19NDD253 pKa = 3.31YY254 pKa = 10.96QQALKK259 pKa = 9.42TVQEE263 pKa = 4.35GLSVDD268 pKa = 3.66QYY270 pKa = 12.02NEE272 pKa = 3.55NLYY275 pKa = 10.78AQAADD280 pKa = 3.96YY281 pKa = 10.86ASHH284 pKa = 7.13LGDD287 pKa = 5.36DD288 pKa = 4.31PLMDD292 pKa = 3.41QYY294 pKa = 11.39LQRR297 pKa = 11.84AHH299 pKa = 7.24EE300 pKa = 4.45IDD302 pKa = 4.37PDD304 pKa = 3.66NLTITIQYY312 pKa = 11.13SNFLLSQQRR321 pKa = 11.84DD322 pKa = 3.44QDD324 pKa = 4.0NLNLLASVEE333 pKa = 4.1DD334 pKa = 4.53DD335 pKa = 3.54EE336 pKa = 6.03VVDD339 pKa = 5.68PQVEE343 pKa = 4.16WNRR346 pKa = 11.84AQSYY350 pKa = 7.82WRR352 pKa = 11.84LEE354 pKa = 3.98QFDD357 pKa = 3.63QAGQAFSAALPAFEE371 pKa = 5.86DD372 pKa = 3.76NPDD375 pKa = 3.95FLHH378 pKa = 6.77QLIDD382 pKa = 3.5YY383 pKa = 7.54YY384 pKa = 10.92QAVGEE389 pKa = 4.2RR390 pKa = 11.84DD391 pKa = 3.44LMFDD395 pKa = 2.93QLQRR399 pKa = 11.84YY400 pKa = 8.44VEE402 pKa = 4.19LVPTDD407 pKa = 3.23TEE409 pKa = 4.03MAEE412 pKa = 3.98RR413 pKa = 11.84LAEE416 pKa = 4.88FEE418 pKa = 4.26DD419 pKa = 3.83QMYY422 pKa = 10.91
Molecular weight: 48.22 kDa
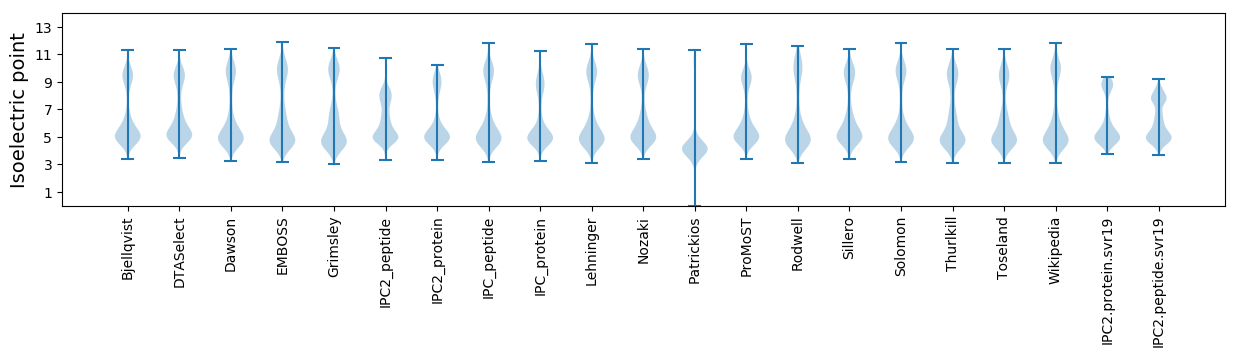
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H4GIE2|H4GIE2_9LACO Uncharacterized protein OS=Lactobacillus gastricus PS3 OX=1144300 GN=PS3_17351 PE=4 SV=1
MM1 pKa = 7.92KK2 pKa = 10.14KK3 pKa = 9.69RR4 pKa = 11.84QYY6 pKa = 11.11SRR8 pKa = 11.84WLVLGIFLLAACMRR22 pKa = 11.84SPFTSLPSVIDD33 pKa = 3.39QIAASFHH40 pKa = 5.82QPATNLGILTTIPLICFGLCSTIVPSISRR69 pKa = 11.84RR70 pKa = 11.84WGNEE74 pKa = 3.11VTLGVALLIMVIGSFLRR91 pKa = 11.84IFNFVTLIFGTILVGIAITFLNVLLPAVITEE122 pKa = 5.09KK123 pKa = 10.37IPQKK127 pKa = 10.45IGLLTGMYY135 pKa = 9.5NVSLAIFSAIGAYY148 pKa = 10.27VITPVTQATNWQTAIIGFTLLIGVTFIVWLPNLRR182 pKa = 11.84FNHH185 pKa = 5.73RR186 pKa = 11.84HH187 pKa = 5.41LVPIQAGNQTRR198 pKa = 11.84IWTNPRR204 pKa = 11.84AWLLLMYY211 pKa = 10.67FGLQSFAFYY220 pKa = 10.24IIVAWLPSIAMAAGLSNATASLVASSFQLLSLPAAFLIPTITPRR264 pKa = 11.84FKK266 pKa = 11.02NRR268 pKa = 11.84VPLVIMAATPLLIGLILLLWPLNSLAYY295 pKa = 10.06FIFISFLLGTGTSATFGIVITLFGLKK321 pKa = 7.84THH323 pKa = 6.27SPEE326 pKa = 3.8EE327 pKa = 4.04TSRR330 pKa = 11.84LSGMVQSVGYY340 pKa = 9.65ILAALGPVIAGNIKK354 pKa = 10.21AATGNWQASLWMTIIIVVIMIAFGALSEE382 pKa = 4.02RR383 pKa = 11.84HH384 pKa = 5.3QYY386 pKa = 10.28INN388 pKa = 3.43
MM1 pKa = 7.92KK2 pKa = 10.14KK3 pKa = 9.69RR4 pKa = 11.84QYY6 pKa = 11.11SRR8 pKa = 11.84WLVLGIFLLAACMRR22 pKa = 11.84SPFTSLPSVIDD33 pKa = 3.39QIAASFHH40 pKa = 5.82QPATNLGILTTIPLICFGLCSTIVPSISRR69 pKa = 11.84RR70 pKa = 11.84WGNEE74 pKa = 3.11VTLGVALLIMVIGSFLRR91 pKa = 11.84IFNFVTLIFGTILVGIAITFLNVLLPAVITEE122 pKa = 5.09KK123 pKa = 10.37IPQKK127 pKa = 10.45IGLLTGMYY135 pKa = 9.5NVSLAIFSAIGAYY148 pKa = 10.27VITPVTQATNWQTAIIGFTLLIGVTFIVWLPNLRR182 pKa = 11.84FNHH185 pKa = 5.73RR186 pKa = 11.84HH187 pKa = 5.41LVPIQAGNQTRR198 pKa = 11.84IWTNPRR204 pKa = 11.84AWLLLMYY211 pKa = 10.67FGLQSFAFYY220 pKa = 10.24IIVAWLPSIAMAAGLSNATASLVASSFQLLSLPAAFLIPTITPRR264 pKa = 11.84FKK266 pKa = 11.02NRR268 pKa = 11.84VPLVIMAATPLLIGLILLLWPLNSLAYY295 pKa = 10.06FIFISFLLGTGTSATFGIVITLFGLKK321 pKa = 7.84THH323 pKa = 6.27SPEE326 pKa = 3.8EE327 pKa = 4.04TSRR330 pKa = 11.84LSGMVQSVGYY340 pKa = 9.65ILAALGPVIAGNIKK354 pKa = 10.21AATGNWQASLWMTIIIVVIMIAFGALSEE382 pKa = 4.02RR383 pKa = 11.84HH384 pKa = 5.3QYY386 pKa = 10.28INN388 pKa = 3.43
Molecular weight: 42.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
425951 |
68 |
1444 |
335.7 |
37.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.348 ± 0.061 | 0.516 ± 0.014 |
6.048 ± 0.066 | 5.548 ± 0.073 |
4.001 ± 0.052 | 6.915 ± 0.05 |
2.316 ± 0.027 | 6.96 ± 0.064 |
5.28 ± 0.058 | 9.747 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.799 ± 0.03 | 4.458 ± 0.041 |
3.791 ± 0.033 | 5.488 ± 0.067 |
4.258 ± 0.049 | 5.542 ± 0.058 |
5.97 ± 0.047 | 7.329 ± 0.052 |
1.102 ± 0.028 | 3.584 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
