
Capsicum baccatum (Peruvian pepper)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Solanoideae; Capsiceae; Capsicum
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 35641 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
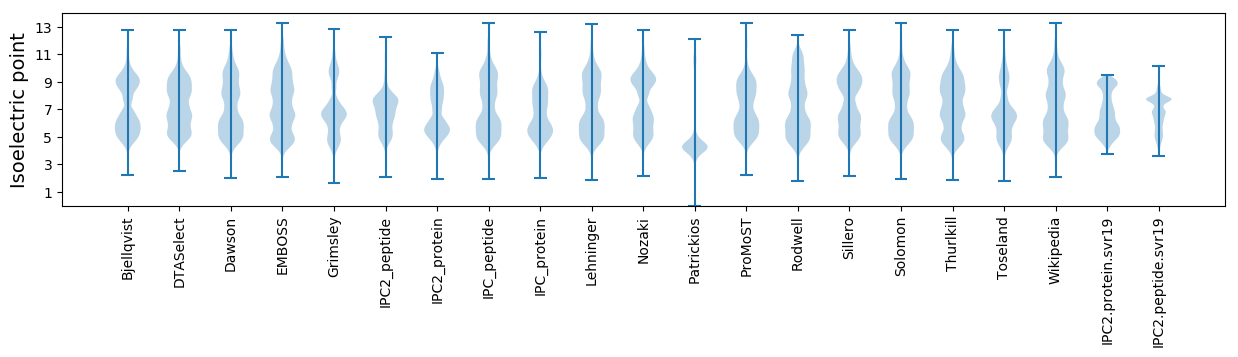
Summary statistics related to proteome-wise predictions
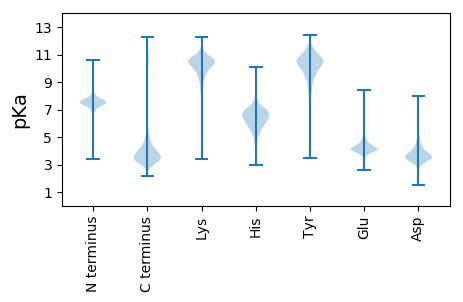
Protein with the lowest isoelectric point:
>tr|A0A2G2V1C7|A0A2G2V1C7_CAPBA Serine/threonine-protein kinase TOR OS=Capsicum baccatum OX=33114 GN=CQW23_33612 PE=4 SV=1
MM1 pKa = 7.08NGEE4 pKa = 4.11VRR6 pKa = 11.84WYY8 pKa = 9.31VSSRR12 pKa = 11.84FSFTLSLVGLVFIIDD27 pKa = 4.97CITYY31 pKa = 8.67ICVDD35 pKa = 3.2GVMVLDD41 pKa = 4.02EE42 pKa = 4.52FEE44 pKa = 5.0GYY46 pKa = 10.17INFMMMPDD54 pKa = 3.57KK55 pKa = 11.14SRR57 pKa = 11.84GYY59 pKa = 8.92MDD61 pKa = 5.71DD62 pKa = 2.92MMMPGDD68 pKa = 3.82SKK70 pKa = 11.5GYY72 pKa = 7.76TDD74 pKa = 5.23VMMVPNLFGGYY85 pKa = 9.14NDD87 pKa = 5.33DD88 pKa = 5.45MMMPDD93 pKa = 3.44NFVGYY98 pKa = 10.22VDD100 pKa = 6.49DD101 pKa = 5.73IMLPGNSKK109 pKa = 10.88GYY111 pKa = 9.76IDD113 pKa = 6.05DD114 pKa = 4.17VMIPDD119 pKa = 3.81KK120 pKa = 11.1FKK122 pKa = 11.06GYY124 pKa = 9.09TDD126 pKa = 5.02DD127 pKa = 5.77IYY129 pKa = 11.57VVWDD133 pKa = 3.61AVMDD137 pKa = 3.93SLLYY141 pKa = 10.65VLLGVLPDD149 pKa = 3.97GMGHH153 pKa = 6.96RR154 pKa = 11.84GNFGQRR160 pKa = 11.84AIQPKK165 pKa = 9.76
MM1 pKa = 7.08NGEE4 pKa = 4.11VRR6 pKa = 11.84WYY8 pKa = 9.31VSSRR12 pKa = 11.84FSFTLSLVGLVFIIDD27 pKa = 4.97CITYY31 pKa = 8.67ICVDD35 pKa = 3.2GVMVLDD41 pKa = 4.02EE42 pKa = 4.52FEE44 pKa = 5.0GYY46 pKa = 10.17INFMMMPDD54 pKa = 3.57KK55 pKa = 11.14SRR57 pKa = 11.84GYY59 pKa = 8.92MDD61 pKa = 5.71DD62 pKa = 2.92MMMPGDD68 pKa = 3.82SKK70 pKa = 11.5GYY72 pKa = 7.76TDD74 pKa = 5.23VMMVPNLFGGYY85 pKa = 9.14NDD87 pKa = 5.33DD88 pKa = 5.45MMMPDD93 pKa = 3.44NFVGYY98 pKa = 10.22VDD100 pKa = 6.49DD101 pKa = 5.73IMLPGNSKK109 pKa = 10.88GYY111 pKa = 9.76IDD113 pKa = 6.05DD114 pKa = 4.17VMIPDD119 pKa = 3.81KK120 pKa = 11.1FKK122 pKa = 11.06GYY124 pKa = 9.09TDD126 pKa = 5.02DD127 pKa = 5.77IYY129 pKa = 11.57VVWDD133 pKa = 3.61AVMDD137 pKa = 3.93SLLYY141 pKa = 10.65VLLGVLPDD149 pKa = 3.97GMGHH153 pKa = 6.96RR154 pKa = 11.84GNFGQRR160 pKa = 11.84AIQPKK165 pKa = 9.76
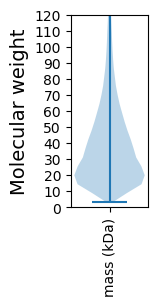
Molecular weight: 18.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G2V2L8|A0A2G2V2L8_CAPBA Ethylene-responsive transcription factor 4 OS=Capsicum baccatum OX=33114 GN=CQW23_33169 PE=4 SV=1
MM1 pKa = 7.65APGHH5 pKa = 6.79HH6 pKa = 6.09GTKK9 pKa = 10.36APWRR13 pKa = 11.84HH14 pKa = 4.83ATVAPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GTMASGHH29 pKa = 6.54KK30 pKa = 9.1GTMTARR36 pKa = 11.84HH37 pKa = 5.91QGTVAPWHH45 pKa = 6.82RR46 pKa = 11.84GTKK49 pKa = 9.38AAWHH53 pKa = 6.6HH54 pKa = 5.22RR55 pKa = 11.84TKK57 pKa = 11.05APWHH61 pKa = 4.92QVGVQPWHH69 pKa = 6.38QGTMPPRR76 pKa = 11.84HH77 pKa = 6.81HH78 pKa = 7.17DD79 pKa = 3.24TVAPRR84 pKa = 11.84RR85 pKa = 11.84PCTLAPRR92 pKa = 11.84HH93 pKa = 6.28RR94 pKa = 11.84GTKK97 pKa = 8.84VAWHH101 pKa = 7.05HH102 pKa = 5.45GTVAPWHH109 pKa = 5.73QGTRR113 pKa = 11.84AAWHH117 pKa = 6.66HH118 pKa = 5.93GAMAPGHH125 pKa = 6.09QGTVAPRR132 pKa = 11.84WRR134 pKa = 11.84GTMAHH139 pKa = 6.28RR140 pKa = 11.84HH141 pKa = 5.91HH142 pKa = 6.59GTVAPWNPGTMAPWHH157 pKa = 6.76HH158 pKa = 6.14GTNASRR164 pKa = 11.84HH165 pKa = 5.12QGTVTPWHH173 pKa = 7.02HH174 pKa = 5.61GTVAPWHH181 pKa = 6.44RR182 pKa = 11.84GTRR185 pKa = 11.84ALRR188 pKa = 11.84HH189 pKa = 6.31HH190 pKa = 6.42GTMAPWHH197 pKa = 7.0HH198 pKa = 5.86GTMAPRR204 pKa = 11.84HH205 pKa = 6.08RR206 pKa = 11.84GLVAAA211 pKa = 5.66
MM1 pKa = 7.65APGHH5 pKa = 6.79HH6 pKa = 6.09GTKK9 pKa = 10.36APWRR13 pKa = 11.84HH14 pKa = 4.83ATVAPRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GTMASGHH29 pKa = 6.54KK30 pKa = 9.1GTMTARR36 pKa = 11.84HH37 pKa = 5.91QGTVAPWHH45 pKa = 6.82RR46 pKa = 11.84GTKK49 pKa = 9.38AAWHH53 pKa = 6.6HH54 pKa = 5.22RR55 pKa = 11.84TKK57 pKa = 11.05APWHH61 pKa = 4.92QVGVQPWHH69 pKa = 6.38QGTMPPRR76 pKa = 11.84HH77 pKa = 6.81HH78 pKa = 7.17DD79 pKa = 3.24TVAPRR84 pKa = 11.84RR85 pKa = 11.84PCTLAPRR92 pKa = 11.84HH93 pKa = 6.28RR94 pKa = 11.84GTKK97 pKa = 8.84VAWHH101 pKa = 7.05HH102 pKa = 5.45GTVAPWHH109 pKa = 5.73QGTRR113 pKa = 11.84AAWHH117 pKa = 6.66HH118 pKa = 5.93GAMAPGHH125 pKa = 6.09QGTVAPRR132 pKa = 11.84WRR134 pKa = 11.84GTMAHH139 pKa = 6.28RR140 pKa = 11.84HH141 pKa = 5.91HH142 pKa = 6.59GTVAPWNPGTMAPWHH157 pKa = 6.76HH158 pKa = 6.14GTNASRR164 pKa = 11.84HH165 pKa = 5.12QGTVTPWHH173 pKa = 7.02HH174 pKa = 5.61GTVAPWHH181 pKa = 6.44RR182 pKa = 11.84GTRR185 pKa = 11.84ALRR188 pKa = 11.84HH189 pKa = 6.31HH190 pKa = 6.42GTMAPWHH197 pKa = 7.0HH198 pKa = 5.86GTMAPRR204 pKa = 11.84HH205 pKa = 6.08RR206 pKa = 11.84GLVAAA211 pKa = 5.66
Molecular weight: 23.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13376091 |
28 |
5007 |
375.3 |
42.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.355 ± 0.012 | 1.907 ± 0.007 |
5.398 ± 0.011 | 6.437 ± 0.013 |
4.201 ± 0.008 | 6.369 ± 0.012 |
2.387 ± 0.005 | 5.587 ± 0.01 |
6.303 ± 0.011 | 9.798 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.497 ± 0.006 | 4.644 ± 0.009 |
4.837 ± 0.015 | 3.555 ± 0.008 |
5.232 ± 0.011 | 8.777 ± 0.014 |
4.93 ± 0.008 | 6.554 ± 0.01 |
1.293 ± 0.005 | 2.925 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
