
Anaeromassilibacillus sp. An250
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Anaeromassilibacillus; unclassified Anaeromassilibacillus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
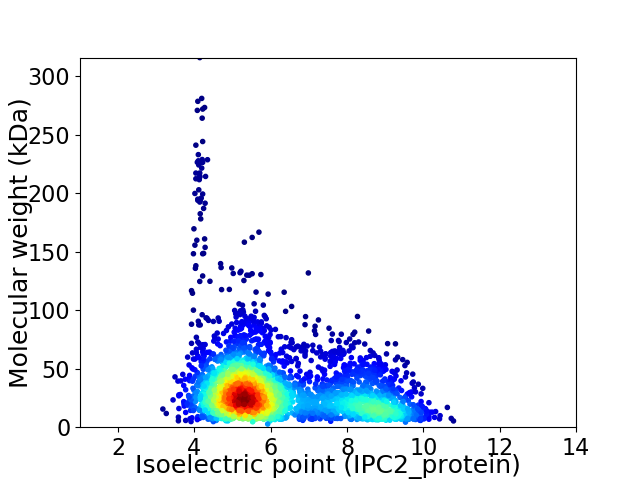
Virtual 2D-PAGE plot for 3281 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4EWC9|A0A1Y4EWC9_9FIRM ATP-dependent DNA helicase RecG OS=Anaeromassilibacillus sp. An250 OX=1965604 GN=recG PE=3 SV=1
MM1 pKa = 7.01KK2 pKa = 9.4TWKK5 pKa = 9.88KK6 pKa = 10.29CLAAVLSLVMMVSATACSGTDD27 pKa = 3.14KK28 pKa = 10.9SWAVKK33 pKa = 10.38SEE35 pKa = 3.83NLTVPIGAYY44 pKa = 9.45IYY46 pKa = 10.31NLYY49 pKa = 10.36SAYY52 pKa = 9.55QQAYY56 pKa = 9.81YY57 pKa = 9.52MVEE60 pKa = 4.33DD61 pKa = 3.56ATQPVLEE68 pKa = 4.44QTVEE72 pKa = 4.12DD73 pKa = 3.66QDD75 pKa = 3.49AATWMKK81 pKa = 11.02DD82 pKa = 3.09FALRR86 pKa = 11.84QTKK89 pKa = 10.25SILVIDD95 pKa = 4.45DD96 pKa = 3.4MMRR99 pKa = 11.84DD100 pKa = 3.36MGLSLTEE107 pKa = 5.49DD108 pKa = 3.77EE109 pKa = 4.82LQQVSDD115 pKa = 3.74QTDD118 pKa = 3.77SFWGSISTAMTEE130 pKa = 3.99YY131 pKa = 10.92GVAKK135 pKa = 10.48SSFNLAYY142 pKa = 10.24ADD144 pKa = 4.37YY145 pKa = 8.33YY146 pKa = 11.1TKK148 pKa = 9.89FTKK151 pKa = 10.46VFDD154 pKa = 3.59ATYY157 pKa = 10.35GPGGTEE163 pKa = 3.79EE164 pKa = 4.81VSDD167 pKa = 4.44EE168 pKa = 4.15DD169 pKa = 4.0VKK171 pKa = 11.28DD172 pKa = 3.98YY173 pKa = 11.67YY174 pKa = 9.93LTNYY178 pKa = 9.86KK179 pKa = 10.41AFSYY183 pKa = 11.21VMAPIIDD190 pKa = 4.06MTTGASLSDD199 pKa = 4.41DD200 pKa = 4.09EE201 pKa = 5.99LSQLDD206 pKa = 4.2TEE208 pKa = 4.75FKK210 pKa = 10.83GYY212 pKa = 10.9ASQINDD218 pKa = 3.09GSMTIQDD225 pKa = 3.53AADD228 pKa = 3.77AYY230 pKa = 9.68KK231 pKa = 10.68ASSGDD236 pKa = 3.42DD237 pKa = 3.33TVQVYY242 pKa = 9.45TDD244 pKa = 3.36AADD247 pKa = 4.08FSGEE251 pKa = 4.07TASYY255 pKa = 9.07PLEE258 pKa = 4.72FGTEE262 pKa = 4.06LDD264 pKa = 3.46TLEE267 pKa = 4.29NGKK270 pKa = 10.62ASVIDD275 pKa = 3.59VQGLYY280 pKa = 10.33KK281 pKa = 10.58VLLVKK286 pKa = 10.78DD287 pKa = 5.16DD288 pKa = 4.06INATADD294 pKa = 3.46EE295 pKa = 5.09RR296 pKa = 11.84ISDD299 pKa = 3.77EE300 pKa = 4.07TSRR303 pKa = 11.84TSILQTMKK311 pKa = 10.63QEE313 pKa = 3.93EE314 pKa = 4.47FQAKK318 pKa = 9.44IDD320 pKa = 4.26DD321 pKa = 4.49AADD324 pKa = 3.64SYY326 pKa = 11.76NATVNQSAIDD336 pKa = 3.81SYY338 pKa = 11.19QPDD341 pKa = 3.63MFVQEE346 pKa = 4.41TSSVAASTDD355 pKa = 3.43EE356 pKa = 4.22TSSQEE361 pKa = 4.06GTDD364 pKa = 3.6GSGEE368 pKa = 4.18EE369 pKa = 4.45TSSEE373 pKa = 4.05AAA375 pKa = 3.63
MM1 pKa = 7.01KK2 pKa = 9.4TWKK5 pKa = 9.88KK6 pKa = 10.29CLAAVLSLVMMVSATACSGTDD27 pKa = 3.14KK28 pKa = 10.9SWAVKK33 pKa = 10.38SEE35 pKa = 3.83NLTVPIGAYY44 pKa = 9.45IYY46 pKa = 10.31NLYY49 pKa = 10.36SAYY52 pKa = 9.55QQAYY56 pKa = 9.81YY57 pKa = 9.52MVEE60 pKa = 4.33DD61 pKa = 3.56ATQPVLEE68 pKa = 4.44QTVEE72 pKa = 4.12DD73 pKa = 3.66QDD75 pKa = 3.49AATWMKK81 pKa = 11.02DD82 pKa = 3.09FALRR86 pKa = 11.84QTKK89 pKa = 10.25SILVIDD95 pKa = 4.45DD96 pKa = 3.4MMRR99 pKa = 11.84DD100 pKa = 3.36MGLSLTEE107 pKa = 5.49DD108 pKa = 3.77EE109 pKa = 4.82LQQVSDD115 pKa = 3.74QTDD118 pKa = 3.77SFWGSISTAMTEE130 pKa = 3.99YY131 pKa = 10.92GVAKK135 pKa = 10.48SSFNLAYY142 pKa = 10.24ADD144 pKa = 4.37YY145 pKa = 8.33YY146 pKa = 11.1TKK148 pKa = 9.89FTKK151 pKa = 10.46VFDD154 pKa = 3.59ATYY157 pKa = 10.35GPGGTEE163 pKa = 3.79EE164 pKa = 4.81VSDD167 pKa = 4.44EE168 pKa = 4.15DD169 pKa = 4.0VKK171 pKa = 11.28DD172 pKa = 3.98YY173 pKa = 11.67YY174 pKa = 9.93LTNYY178 pKa = 9.86KK179 pKa = 10.41AFSYY183 pKa = 11.21VMAPIIDD190 pKa = 4.06MTTGASLSDD199 pKa = 4.41DD200 pKa = 4.09EE201 pKa = 5.99LSQLDD206 pKa = 4.2TEE208 pKa = 4.75FKK210 pKa = 10.83GYY212 pKa = 10.9ASQINDD218 pKa = 3.09GSMTIQDD225 pKa = 3.53AADD228 pKa = 3.77AYY230 pKa = 9.68KK231 pKa = 10.68ASSGDD236 pKa = 3.42DD237 pKa = 3.33TVQVYY242 pKa = 9.45TDD244 pKa = 3.36AADD247 pKa = 4.08FSGEE251 pKa = 4.07TASYY255 pKa = 9.07PLEE258 pKa = 4.72FGTEE262 pKa = 4.06LDD264 pKa = 3.46TLEE267 pKa = 4.29NGKK270 pKa = 10.62ASVIDD275 pKa = 3.59VQGLYY280 pKa = 10.33KK281 pKa = 10.58VLLVKK286 pKa = 10.78DD287 pKa = 5.16DD288 pKa = 4.06INATADD294 pKa = 3.46EE295 pKa = 5.09RR296 pKa = 11.84ISDD299 pKa = 3.77EE300 pKa = 4.07TSRR303 pKa = 11.84TSILQTMKK311 pKa = 10.63QEE313 pKa = 3.93EE314 pKa = 4.47FQAKK318 pKa = 9.44IDD320 pKa = 4.26DD321 pKa = 4.49AADD324 pKa = 3.64SYY326 pKa = 11.76NATVNQSAIDD336 pKa = 3.81SYY338 pKa = 11.19QPDD341 pKa = 3.63MFVQEE346 pKa = 4.41TSSVAASTDD355 pKa = 3.43EE356 pKa = 4.22TSSQEE361 pKa = 4.06GTDD364 pKa = 3.6GSGEE368 pKa = 4.18EE369 pKa = 4.45TSSEE373 pKa = 4.05AAA375 pKa = 3.63
Molecular weight: 41.06 kDa
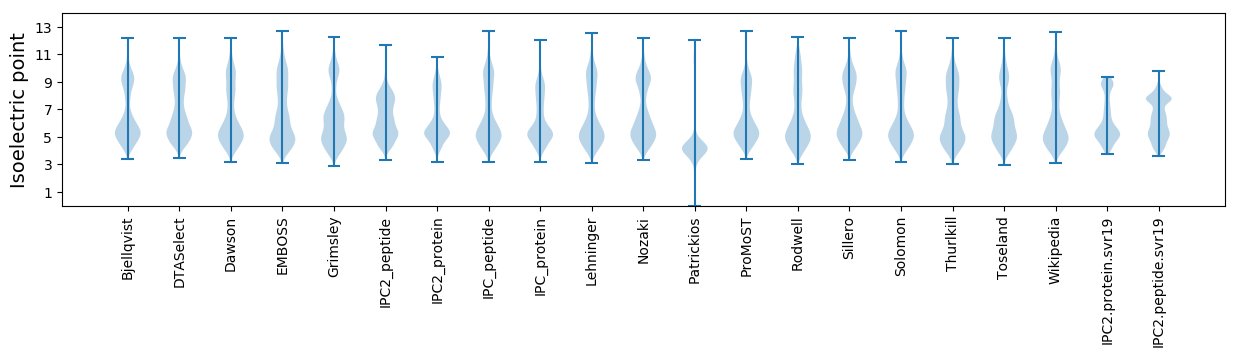
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4ENZ6|A0A1Y4ENZ6_9FIRM LacI family transcriptional regulator OS=Anaeromassilibacillus sp. An250 OX=1965604 GN=B5F54_12440 PE=4 SV=1
MM1 pKa = 7.49TSFTNGVEE9 pKa = 4.66LIDD12 pKa = 4.12PRR14 pKa = 11.84CLLGKK19 pKa = 9.78HH20 pKa = 5.59EE21 pKa = 4.59KK22 pKa = 9.83AAPAIGCRR30 pKa = 11.84PFAFLHH36 pKa = 6.43IGTICLEE43 pKa = 3.77VRR45 pKa = 11.84IRR47 pKa = 11.84SAALIFLARR56 pKa = 11.84HH57 pKa = 4.78KK58 pKa = 9.45TAILGATLLAGVVRR72 pKa = 11.84QLRR75 pKa = 11.84GTGCQPVHH83 pKa = 6.52SRR85 pKa = 11.84IVAVGWQVPTCGVGVDD101 pKa = 3.72RR102 pKa = 11.84SSSVLEE108 pKa = 3.98KK109 pKa = 10.02SWRR112 pKa = 11.84TRR114 pKa = 11.84RR115 pKa = 11.84PAPRR119 pKa = 11.84TMPGAAAIQLWRR131 pKa = 11.84GSTPLIYY138 pKa = 9.89QGHH141 pKa = 6.69LEE143 pKa = 4.35DD144 pKa = 4.79VFF146 pKa = 4.87
MM1 pKa = 7.49TSFTNGVEE9 pKa = 4.66LIDD12 pKa = 4.12PRR14 pKa = 11.84CLLGKK19 pKa = 9.78HH20 pKa = 5.59EE21 pKa = 4.59KK22 pKa = 9.83AAPAIGCRR30 pKa = 11.84PFAFLHH36 pKa = 6.43IGTICLEE43 pKa = 3.77VRR45 pKa = 11.84IRR47 pKa = 11.84SAALIFLARR56 pKa = 11.84HH57 pKa = 4.78KK58 pKa = 9.45TAILGATLLAGVVRR72 pKa = 11.84QLRR75 pKa = 11.84GTGCQPVHH83 pKa = 6.52SRR85 pKa = 11.84IVAVGWQVPTCGVGVDD101 pKa = 3.72RR102 pKa = 11.84SSSVLEE108 pKa = 3.98KK109 pKa = 10.02SWRR112 pKa = 11.84TRR114 pKa = 11.84RR115 pKa = 11.84PAPRR119 pKa = 11.84TMPGAAAIQLWRR131 pKa = 11.84GSTPLIYY138 pKa = 9.89QGHH141 pKa = 6.69LEE143 pKa = 4.35DD144 pKa = 4.79VFF146 pKa = 4.87
Molecular weight: 15.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1017378 |
27 |
2863 |
310.1 |
34.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.999 ± 0.05 | 1.554 ± 0.021 |
5.715 ± 0.045 | 7.257 ± 0.044 |
3.918 ± 0.031 | 7.335 ± 0.035 |
1.835 ± 0.02 | 6.001 ± 0.043 |
5.336 ± 0.042 | 9.349 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.725 ± 0.018 | 3.831 ± 0.035 |
4.063 ± 0.03 | 3.771 ± 0.025 |
5.248 ± 0.054 | 5.777 ± 0.034 |
5.574 ± 0.051 | 6.959 ± 0.039 |
1.026 ± 0.015 | 3.728 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
