
Human associated huchismacovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Huchismacovirus
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
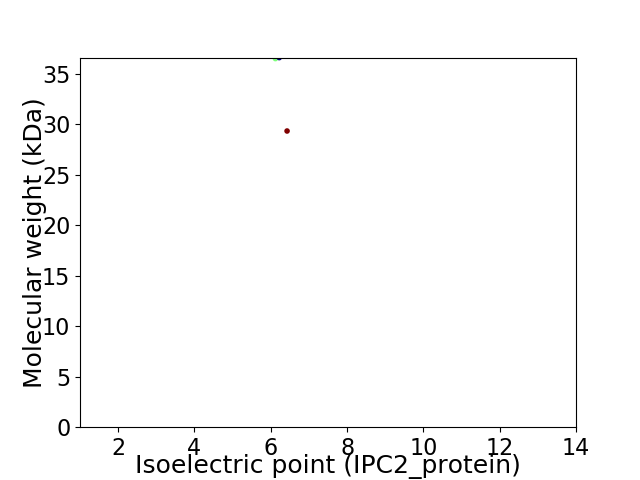
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
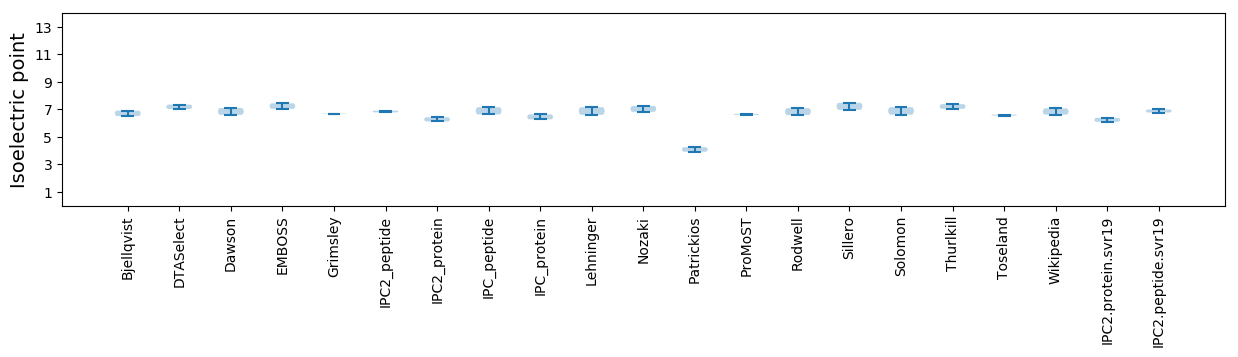
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5GG74|A0A0B5GG74_9VIRU Rep protein OS=Human associated huchismacovirus 3 OX=2169936 GN=rep PE=4 SV=1
MM1 pKa = 6.3MQKK4 pKa = 9.97FRR6 pKa = 11.84WFVDD10 pKa = 4.0LSTTPEE16 pKa = 3.89RR17 pKa = 11.84VSVLKK22 pKa = 9.05ITCGGTRR29 pKa = 11.84CLRR32 pKa = 11.84RR33 pKa = 11.84LLPFFLVLISIFKK46 pKa = 10.3LGKK49 pKa = 8.25TSVKK53 pKa = 9.9FVPSSTLPVDD63 pKa = 3.64PTGLSYY69 pKa = 10.68EE70 pKa = 4.23AGEE73 pKa = 4.26NTVDD77 pKa = 3.7PRR79 pKa = 11.84DD80 pKa = 3.66MFNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 3.99DD94 pKa = 3.13LADD97 pKa = 3.4VVLNNISTAEE107 pKa = 4.02DD108 pKa = 3.44QNNVYY113 pKa = 10.21YY114 pKa = 11.53AMMLDD119 pKa = 3.52SRR121 pKa = 11.84WYY123 pKa = 10.46KK124 pKa = 10.42FRR126 pKa = 11.84LQDD129 pKa = 3.24GFRR132 pKa = 11.84RR133 pKa = 11.84DD134 pKa = 3.66AVPLYY139 pKa = 9.61WRR141 pKa = 11.84IGQLHH146 pKa = 6.19QDD148 pKa = 3.71NFPGSVNNYY157 pKa = 7.94LTVKK161 pKa = 10.51SDD163 pKa = 3.4VTKK166 pKa = 10.88SPTVSHH172 pKa = 6.99EE173 pKa = 3.56IGYY176 pKa = 9.2YY177 pKa = 10.06AYY179 pKa = 10.27SSPGSASPDD188 pKa = 2.97RR189 pKa = 11.84KK190 pKa = 10.31NQILLGSSPHH200 pKa = 5.59GTFQFGHH207 pKa = 6.67HH208 pKa = 7.19DD209 pKa = 3.93KK210 pKa = 11.0IGWLPTDD217 pKa = 3.76MFTNNYY223 pKa = 9.04VNTSGTPTMSPRR235 pKa = 11.84TPPEE239 pKa = 3.59VDD241 pKa = 3.4LLTVVLPAAYY251 pKa = 7.44KK252 pKa = 8.85TRR254 pKa = 11.84FYY256 pKa = 10.76YY257 pKa = 10.28RR258 pKa = 11.84LYY260 pKa = 9.16ITEE263 pKa = 3.9EE264 pKa = 4.72VYY266 pKa = 10.79FKK268 pKa = 11.13EE269 pKa = 4.21PVSIFAGQLSDD280 pKa = 3.41GQYY283 pKa = 11.18VSTTMDD289 pKa = 3.11RR290 pKa = 11.84FTYY293 pKa = 7.8PTLCGKK299 pKa = 10.16FPGNLNGVPSGTFLPVEE316 pKa = 4.61LDD318 pKa = 3.27NSGGDD323 pKa = 3.59TPP325 pKa = 5.24
MM1 pKa = 6.3MQKK4 pKa = 9.97FRR6 pKa = 11.84WFVDD10 pKa = 4.0LSTTPEE16 pKa = 3.89RR17 pKa = 11.84VSVLKK22 pKa = 9.05ITCGGTRR29 pKa = 11.84CLRR32 pKa = 11.84RR33 pKa = 11.84LLPFFLVLISIFKK46 pKa = 10.3LGKK49 pKa = 8.25TSVKK53 pKa = 9.9FVPSSTLPVDD63 pKa = 3.64PTGLSYY69 pKa = 10.68EE70 pKa = 4.23AGEE73 pKa = 4.26NTVDD77 pKa = 3.7PRR79 pKa = 11.84DD80 pKa = 3.66MFNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 3.99DD94 pKa = 3.13LADD97 pKa = 3.4VVLNNISTAEE107 pKa = 4.02DD108 pKa = 3.44QNNVYY113 pKa = 10.21YY114 pKa = 11.53AMMLDD119 pKa = 3.52SRR121 pKa = 11.84WYY123 pKa = 10.46KK124 pKa = 10.42FRR126 pKa = 11.84LQDD129 pKa = 3.24GFRR132 pKa = 11.84RR133 pKa = 11.84DD134 pKa = 3.66AVPLYY139 pKa = 9.61WRR141 pKa = 11.84IGQLHH146 pKa = 6.19QDD148 pKa = 3.71NFPGSVNNYY157 pKa = 7.94LTVKK161 pKa = 10.51SDD163 pKa = 3.4VTKK166 pKa = 10.88SPTVSHH172 pKa = 6.99EE173 pKa = 3.56IGYY176 pKa = 9.2YY177 pKa = 10.06AYY179 pKa = 10.27SSPGSASPDD188 pKa = 2.97RR189 pKa = 11.84KK190 pKa = 10.31NQILLGSSPHH200 pKa = 5.59GTFQFGHH207 pKa = 6.67HH208 pKa = 7.19DD209 pKa = 3.93KK210 pKa = 11.0IGWLPTDD217 pKa = 3.76MFTNNYY223 pKa = 9.04VNTSGTPTMSPRR235 pKa = 11.84TPPEE239 pKa = 3.59VDD241 pKa = 3.4LLTVVLPAAYY251 pKa = 7.44KK252 pKa = 8.85TRR254 pKa = 11.84FYY256 pKa = 10.76YY257 pKa = 10.28RR258 pKa = 11.84LYY260 pKa = 9.16ITEE263 pKa = 3.9EE264 pKa = 4.72VYY266 pKa = 10.79FKK268 pKa = 11.13EE269 pKa = 4.21PVSIFAGQLSDD280 pKa = 3.41GQYY283 pKa = 11.18VSTTMDD289 pKa = 3.11RR290 pKa = 11.84FTYY293 pKa = 7.8PTLCGKK299 pKa = 10.16FPGNLNGVPSGTFLPVEE316 pKa = 4.61LDD318 pKa = 3.27NSGGDD323 pKa = 3.59TPP325 pKa = 5.24
Molecular weight: 36.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5GG74|A0A0B5GG74_9VIRU Rep protein OS=Human associated huchismacovirus 3 OX=2169936 GN=rep PE=4 SV=1
MM1 pKa = 8.0PSPKK5 pKa = 9.58WYY7 pKa = 9.93DD8 pKa = 2.92ITASKK13 pKa = 10.69EE14 pKa = 3.7KK15 pKa = 10.42LPRR18 pKa = 11.84SILEE22 pKa = 3.43KK23 pKa = 9.95WLNEE27 pKa = 3.72RR28 pKa = 11.84CEE30 pKa = 3.58RR31 pKa = 11.84WAYY34 pKa = 9.79GNEE37 pKa = 4.06VGEE40 pKa = 4.6GGYY43 pKa = 8.44EE44 pKa = 3.75HH45 pKa = 6.4YY46 pKa = 10.46QIRR49 pKa = 11.84VVLKK53 pKa = 9.87EE54 pKa = 3.86PTDD57 pKa = 3.66EE58 pKa = 4.3YY59 pKa = 11.41EE60 pKa = 4.06MRR62 pKa = 11.84KK63 pKa = 7.73MWAAFGHH70 pKa = 5.77VSPTHH75 pKa = 5.49VRR77 pKa = 11.84NFDD80 pKa = 3.63YY81 pKa = 10.87VLKK84 pKa = 10.68EE85 pKa = 3.62GDD87 pKa = 4.61YY88 pKa = 8.56VCSWIKK94 pKa = 10.53IPEE97 pKa = 4.04NMQAATFRR105 pKa = 11.84PWQRR109 pKa = 11.84ALLDD113 pKa = 3.89LEE115 pKa = 4.19QDD117 pKa = 3.5DD118 pKa = 4.25RR119 pKa = 11.84QIDD122 pKa = 4.13VIIDD126 pKa = 3.58LAGNTGKK133 pKa = 10.0SWLTKK138 pKa = 9.6YY139 pKa = 10.33CALKK143 pKa = 9.74MRR145 pKa = 11.84AINVPSGLEE154 pKa = 3.86GKK156 pKa = 10.17DD157 pKa = 3.27IMRR160 pKa = 11.84MCLKK164 pKa = 10.52RR165 pKa = 11.84GAQSGLYY172 pKa = 9.5IFDD175 pKa = 3.76MPRR178 pKa = 11.84AITKK182 pKa = 9.11QSKK185 pKa = 9.26SVWSAIEE192 pKa = 4.23SIKK195 pKa = 10.93NGYY198 pKa = 8.87LWDD201 pKa = 4.71DD202 pKa = 3.86RR203 pKa = 11.84YY204 pKa = 8.77TWEE207 pKa = 4.92EE208 pKa = 3.62KK209 pKa = 10.09WIDD212 pKa = 3.66PPRR215 pKa = 11.84IWVFTNEE222 pKa = 3.93YY223 pKa = 9.29PKK225 pKa = 10.9YY226 pKa = 10.45DD227 pKa = 3.78LVSEE231 pKa = 4.11DD232 pKa = 4.79RR233 pKa = 11.84LRR235 pKa = 11.84FWAPTPTGLVSLAHH249 pKa = 6.38
MM1 pKa = 8.0PSPKK5 pKa = 9.58WYY7 pKa = 9.93DD8 pKa = 2.92ITASKK13 pKa = 10.69EE14 pKa = 3.7KK15 pKa = 10.42LPRR18 pKa = 11.84SILEE22 pKa = 3.43KK23 pKa = 9.95WLNEE27 pKa = 3.72RR28 pKa = 11.84CEE30 pKa = 3.58RR31 pKa = 11.84WAYY34 pKa = 9.79GNEE37 pKa = 4.06VGEE40 pKa = 4.6GGYY43 pKa = 8.44EE44 pKa = 3.75HH45 pKa = 6.4YY46 pKa = 10.46QIRR49 pKa = 11.84VVLKK53 pKa = 9.87EE54 pKa = 3.86PTDD57 pKa = 3.66EE58 pKa = 4.3YY59 pKa = 11.41EE60 pKa = 4.06MRR62 pKa = 11.84KK63 pKa = 7.73MWAAFGHH70 pKa = 5.77VSPTHH75 pKa = 5.49VRR77 pKa = 11.84NFDD80 pKa = 3.63YY81 pKa = 10.87VLKK84 pKa = 10.68EE85 pKa = 3.62GDD87 pKa = 4.61YY88 pKa = 8.56VCSWIKK94 pKa = 10.53IPEE97 pKa = 4.04NMQAATFRR105 pKa = 11.84PWQRR109 pKa = 11.84ALLDD113 pKa = 3.89LEE115 pKa = 4.19QDD117 pKa = 3.5DD118 pKa = 4.25RR119 pKa = 11.84QIDD122 pKa = 4.13VIIDD126 pKa = 3.58LAGNTGKK133 pKa = 10.0SWLTKK138 pKa = 9.6YY139 pKa = 10.33CALKK143 pKa = 9.74MRR145 pKa = 11.84AINVPSGLEE154 pKa = 3.86GKK156 pKa = 10.17DD157 pKa = 3.27IMRR160 pKa = 11.84MCLKK164 pKa = 10.52RR165 pKa = 11.84GAQSGLYY172 pKa = 9.5IFDD175 pKa = 3.76MPRR178 pKa = 11.84AITKK182 pKa = 9.11QSKK185 pKa = 9.26SVWSAIEE192 pKa = 4.23SIKK195 pKa = 10.93NGYY198 pKa = 8.87LWDD201 pKa = 4.71DD202 pKa = 3.86RR203 pKa = 11.84YY204 pKa = 8.77TWEE207 pKa = 4.92EE208 pKa = 3.62KK209 pKa = 10.09WIDD212 pKa = 3.66PPRR215 pKa = 11.84IWVFTNEE222 pKa = 3.93YY223 pKa = 9.29PKK225 pKa = 10.9YY226 pKa = 10.45DD227 pKa = 3.78LVSEE231 pKa = 4.11DD232 pKa = 4.79RR233 pKa = 11.84LRR235 pKa = 11.84FWAPTPTGLVSLAHH249 pKa = 6.38
Molecular weight: 29.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
574 |
249 |
325 |
287.0 |
32.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.355 ± 1.107 | 1.22 ± 0.257 |
6.272 ± 0.102 | 5.226 ± 1.595 |
4.355 ± 1.291 | 6.794 ± 0.777 |
1.568 ± 0.026 | 4.704 ± 1.142 |
5.401 ± 1.213 | 8.537 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.787 ± 0.282 | 4.355 ± 0.758 |
6.794 ± 0.777 | 2.787 ± 0.016 |
5.923 ± 0.6 | 6.969 ± 0.893 |
6.969 ± 1.692 | 6.794 ± 1.044 |
2.962 ± 1.499 | 5.226 ± 0.004 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
