
Streptococcus phage Javan284
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
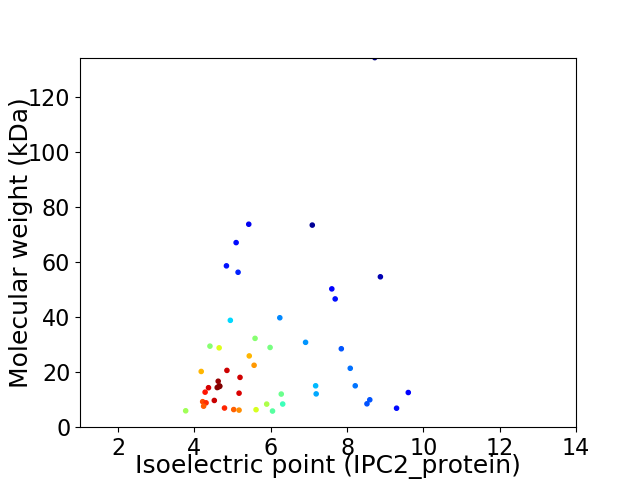
Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4D6AX25|A0A4D6AX25_9CAUD Integrase OS=Streptococcus phage Javan284 OX=2548093 GN=Javan284_0051 PE=4 SV=1
MM1 pKa = 7.31SRR3 pKa = 11.84IEE5 pKa = 3.77TSIAEE10 pKa = 4.26MYY12 pKa = 9.82NLQRR16 pKa = 11.84IPVHH20 pKa = 7.08YY21 pKa = 10.95DD22 pKa = 2.48MGDD25 pKa = 3.2RR26 pKa = 11.84YY27 pKa = 11.26GNDD30 pKa = 3.3ADD32 pKa = 4.42RR33 pKa = 11.84DD34 pKa = 3.87GLIEE38 pKa = 4.2YY39 pKa = 9.11DD40 pKa = 3.73CSSAVSKK47 pKa = 11.09ALGISLSNNTEE58 pKa = 4.09SLKK61 pKa = 11.27SNLPKK66 pKa = 9.93IGYY69 pKa = 8.46NCFYY73 pKa = 10.77DD74 pKa = 4.76GVDD77 pKa = 3.42GTFDD81 pKa = 3.32AVRR84 pKa = 11.84GDD86 pKa = 3.67VVIWGPRR93 pKa = 11.84DD94 pKa = 3.41GLSSLGAFGHH104 pKa = 5.5VLIFVDD110 pKa = 4.75GSNVIHH116 pKa = 6.9CNYY119 pKa = 9.97GSDD122 pKa = 3.26GVTVNDD128 pKa = 3.95YY129 pKa = 11.31NYY131 pKa = 9.83LWRR134 pKa = 11.84INGCPRR140 pKa = 11.84EE141 pKa = 4.18TVFRR145 pKa = 11.84EE146 pKa = 3.52NADD149 pKa = 4.6AINQEE154 pKa = 4.38PKK156 pKa = 9.58PSGRR160 pKa = 11.84KK161 pKa = 8.62IYY163 pKa = 9.98QVNAMEE169 pKa = 4.56FVNGIWQVKK178 pKa = 9.42CDD180 pKa = 3.92YY181 pKa = 10.52LCPVEE186 pKa = 6.06FNWNDD191 pKa = 3.02NGICVGDD198 pKa = 3.68IDD200 pKa = 5.53IVDD203 pKa = 3.65KK204 pKa = 11.28NGNLIADD211 pKa = 4.13QEE213 pKa = 4.87CKK215 pKa = 10.11VGSYY219 pKa = 9.75FVFNPDD225 pKa = 5.3KK226 pKa = 10.77ILSDD230 pKa = 3.21NGGAYY235 pKa = 7.91GTGGYY240 pKa = 7.67YY241 pKa = 9.17WRR243 pKa = 11.84NFTFAEE249 pKa = 4.89GGEE252 pKa = 4.33AWLSTWDD259 pKa = 4.26KK260 pKa = 11.46KK261 pKa = 11.26DD262 pKa = 3.47LLGG265 pKa = 4.92
MM1 pKa = 7.31SRR3 pKa = 11.84IEE5 pKa = 3.77TSIAEE10 pKa = 4.26MYY12 pKa = 9.82NLQRR16 pKa = 11.84IPVHH20 pKa = 7.08YY21 pKa = 10.95DD22 pKa = 2.48MGDD25 pKa = 3.2RR26 pKa = 11.84YY27 pKa = 11.26GNDD30 pKa = 3.3ADD32 pKa = 4.42RR33 pKa = 11.84DD34 pKa = 3.87GLIEE38 pKa = 4.2YY39 pKa = 9.11DD40 pKa = 3.73CSSAVSKK47 pKa = 11.09ALGISLSNNTEE58 pKa = 4.09SLKK61 pKa = 11.27SNLPKK66 pKa = 9.93IGYY69 pKa = 8.46NCFYY73 pKa = 10.77DD74 pKa = 4.76GVDD77 pKa = 3.42GTFDD81 pKa = 3.32AVRR84 pKa = 11.84GDD86 pKa = 3.67VVIWGPRR93 pKa = 11.84DD94 pKa = 3.41GLSSLGAFGHH104 pKa = 5.5VLIFVDD110 pKa = 4.75GSNVIHH116 pKa = 6.9CNYY119 pKa = 9.97GSDD122 pKa = 3.26GVTVNDD128 pKa = 3.95YY129 pKa = 11.31NYY131 pKa = 9.83LWRR134 pKa = 11.84INGCPRR140 pKa = 11.84EE141 pKa = 4.18TVFRR145 pKa = 11.84EE146 pKa = 3.52NADD149 pKa = 4.6AINQEE154 pKa = 4.38PKK156 pKa = 9.58PSGRR160 pKa = 11.84KK161 pKa = 8.62IYY163 pKa = 9.98QVNAMEE169 pKa = 4.56FVNGIWQVKK178 pKa = 9.42CDD180 pKa = 3.92YY181 pKa = 10.52LCPVEE186 pKa = 6.06FNWNDD191 pKa = 3.02NGICVGDD198 pKa = 3.68IDD200 pKa = 5.53IVDD203 pKa = 3.65KK204 pKa = 11.28NGNLIADD211 pKa = 4.13QEE213 pKa = 4.87CKK215 pKa = 10.11VGSYY219 pKa = 9.75FVFNPDD225 pKa = 5.3KK226 pKa = 10.77ILSDD230 pKa = 3.21NGGAYY235 pKa = 7.91GTGGYY240 pKa = 7.67YY241 pKa = 9.17WRR243 pKa = 11.84NFTFAEE249 pKa = 4.89GGEE252 pKa = 4.33AWLSTWDD259 pKa = 4.26KK260 pKa = 11.46KK261 pKa = 11.26DD262 pKa = 3.47LLGG265 pKa = 4.92
Molecular weight: 29.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6AX86|A0A4D6AX86_9CAUD Terminase large subunit OS=Streptococcus phage Javan284 OX=2548093 GN=Javan284_0029 PE=4 SV=1
MM1 pKa = 7.35FRR3 pKa = 11.84IRR5 pKa = 11.84TKK7 pKa = 11.08ADD9 pKa = 3.05LSGAEE14 pKa = 4.38RR15 pKa = 11.84KK16 pKa = 10.24VSDD19 pKa = 3.85ANVLRR24 pKa = 11.84GKK26 pKa = 10.18RR27 pKa = 11.84ALANQVLMDD36 pKa = 3.8TDD38 pKa = 3.96KK39 pKa = 11.12YY40 pKa = 11.24IPMKK44 pKa = 10.69GGALRR49 pKa = 11.84ASGQIAIDD57 pKa = 4.26GSAVSWNTVYY67 pKa = 10.96ARR69 pKa = 11.84AQFYY73 pKa = 7.49GTNGIVVFKK82 pKa = 10.68KK83 pKa = 9.31YY84 pKa = 6.41TTPGTGKK91 pKa = 10.22LWYY94 pKa = 9.87DD95 pKa = 3.77KK96 pKa = 11.01SAEE99 pKa = 4.06ANVDD103 pKa = 2.4KK104 pKa = 10.43WKK106 pKa = 10.65RR107 pKa = 11.84VAAKK111 pKa = 10.65GMGFF115 pKa = 3.41
MM1 pKa = 7.35FRR3 pKa = 11.84IRR5 pKa = 11.84TKK7 pKa = 11.08ADD9 pKa = 3.05LSGAEE14 pKa = 4.38RR15 pKa = 11.84KK16 pKa = 10.24VSDD19 pKa = 3.85ANVLRR24 pKa = 11.84GKK26 pKa = 10.18RR27 pKa = 11.84ALANQVLMDD36 pKa = 3.8TDD38 pKa = 3.96KK39 pKa = 11.12YY40 pKa = 11.24IPMKK44 pKa = 10.69GGALRR49 pKa = 11.84ASGQIAIDD57 pKa = 4.26GSAVSWNTVYY67 pKa = 10.96ARR69 pKa = 11.84AQFYY73 pKa = 7.49GTNGIVVFKK82 pKa = 10.68KK83 pKa = 9.31YY84 pKa = 6.41TTPGTGKK91 pKa = 10.22LWYY94 pKa = 9.87DD95 pKa = 3.77KK96 pKa = 11.01SAEE99 pKa = 4.06ANVDD103 pKa = 2.4KK104 pKa = 10.43WKK106 pKa = 10.65RR107 pKa = 11.84VAAKK111 pKa = 10.65GMGFF115 pKa = 3.41
Molecular weight: 12.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11583 |
49 |
1316 |
227.1 |
25.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.718 ± 1.007 | 0.553 ± 0.111 |
6.432 ± 0.414 | 6.872 ± 0.691 |
4.256 ± 0.194 | 6.553 ± 0.553 |
1.278 ± 0.182 | 6.63 ± 0.328 |
8.02 ± 0.49 | 7.468 ± 0.246 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.22 | 6.233 ± 0.435 |
2.702 ± 0.154 | 4.368 ± 0.366 |
3.591 ± 0.296 | 6.423 ± 0.433 |
6.63 ± 0.545 | 6.423 ± 0.306 |
1.269 ± 0.149 | 4.032 ± 0.392 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
