
Halorubrum pleomorphic virus 6
Taxonomy: Viruses; Monodnaviria; Trapavirae; Saleviricota; Huolimaviricetes; Haloruvirales; Pleolipoviridae; Alphapleolipovirus; Alphapleolipovirus HRPV6
Average proteome isoelectric point is 5.53
Get precalculated fractions of proteins
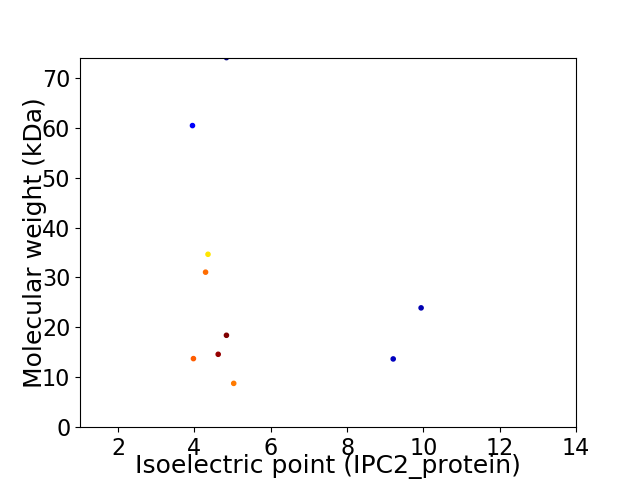
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9ABP7|H9ABP7_9VIRU ORF6 OS=Halorubrum pleomorphic virus 6 OX=1156721 GN=ORF6 PE=4 SV=1
MM1 pKa = 7.55GRR3 pKa = 11.84RR4 pKa = 11.84TFVKK8 pKa = 10.51GLGAAGAASVGLAHH22 pKa = 7.61DD23 pKa = 4.58RR24 pKa = 11.84GAARR28 pKa = 11.84NAEE31 pKa = 4.56AIAPLVGYY39 pKa = 10.29AIGAAAISAVGGIGVGWTLRR59 pKa = 11.84EE60 pKa = 4.03FEE62 pKa = 4.79VVGSDD67 pKa = 4.72DD68 pKa = 3.8PAEE71 pKa = 4.25GLTPDD76 pKa = 3.54VLRR79 pKa = 11.84NQLSDD84 pKa = 3.6SVVKK88 pKa = 10.61RR89 pKa = 11.84KK90 pKa = 10.36SNNQSTMVDD99 pKa = 3.25NQNILDD105 pKa = 4.25GVEE108 pKa = 3.87HH109 pKa = 6.22TAYY112 pKa = 9.83TEE114 pKa = 3.97AKK116 pKa = 8.8IAAIEE121 pKa = 3.95EE122 pKa = 4.43LNAGSSEE129 pKa = 4.28SAVLSAANSAIDD141 pKa = 3.52SYY143 pKa = 10.49EE144 pKa = 3.92TTVRR148 pKa = 11.84TNFYY152 pKa = 10.33KK153 pKa = 10.37SWNEE157 pKa = 3.67TVRR160 pKa = 11.84EE161 pKa = 4.25LEE163 pKa = 4.35AMTQTVIAHH172 pKa = 6.75ADD174 pKa = 3.22VGLSYY179 pKa = 9.8ITDD182 pKa = 3.94FGDD185 pKa = 3.42PRR187 pKa = 11.84FGNLASGTSPNTLKK201 pKa = 10.3DD202 pKa = 3.66TTVSMPDD209 pKa = 3.15GTNFTLLTFRR219 pKa = 11.84HH220 pKa = 4.74NTGWDD225 pKa = 3.43SGNAAYY231 pKa = 10.21SVVEE235 pKa = 4.01YY236 pKa = 10.77NPKK239 pKa = 10.38EE240 pKa = 4.22VVTSTNSNTYY250 pKa = 8.38NTVDD254 pKa = 2.97GTQYY258 pKa = 10.65MKK260 pKa = 10.67FSEE263 pKa = 4.31WNAVEE268 pKa = 4.33TEE270 pKa = 3.86MDD272 pKa = 3.59TVFQNVRR279 pKa = 11.84NGISTWVTNVYY290 pKa = 11.18GDD292 pKa = 3.84VQSGAIEE299 pKa = 4.05ISDD302 pKa = 3.5LVTPRR307 pKa = 11.84EE308 pKa = 4.24RR309 pKa = 11.84ATMMAQEE316 pKa = 4.65EE317 pKa = 4.92GMSQAIADD325 pKa = 5.12LIALNVPVDD334 pKa = 3.84AEE336 pKa = 4.23RR337 pKa = 11.84EE338 pKa = 4.21ATITIQDD345 pKa = 3.56TGATLPGTFALTDD358 pKa = 3.56SSDD361 pKa = 3.78GPLSAGQTYY370 pKa = 10.66DD371 pKa = 3.09PSTFSGDD378 pKa = 3.59VYY380 pKa = 9.75FTADD384 pKa = 3.16MSLVEE389 pKa = 4.75GPWDD393 pKa = 4.62AINSGVDD400 pKa = 2.88GGTITITSEE409 pKa = 4.15PYY411 pKa = 9.2EE412 pKa = 4.25GTAIEE417 pKa = 4.41VTTVEE422 pKa = 4.47SEE424 pKa = 4.64TVSVPAADD432 pKa = 3.35WTDD435 pKa = 3.4NGDD438 pKa = 3.85GTWSYY443 pKa = 11.33DD444 pKa = 3.16ASGDD448 pKa = 4.09LEE450 pKa = 4.42TTITNVDD457 pKa = 3.36SARR460 pKa = 11.84FVSTATEE467 pKa = 4.08TTYY470 pKa = 10.1DD471 pKa = 3.54TLQLKK476 pKa = 10.21GAFTVDD482 pKa = 2.98KK483 pKa = 10.64LVNKK487 pKa = 10.19QSGEE491 pKa = 4.3EE492 pKa = 4.01VSSTSFTSSEE502 pKa = 3.98PQTDD506 pKa = 3.34SNYY509 pKa = 8.7ITQDD513 pKa = 2.37EE514 pKa = 4.42WDD516 pKa = 3.49QLEE519 pKa = 4.23QQNKK523 pKa = 8.28EE524 pKa = 4.17LIEE527 pKa = 4.64KK528 pKa = 10.22YY529 pKa = 9.73EE530 pKa = 3.97QSQSGGGLDD539 pKa = 4.92LGGLDD544 pKa = 3.42MFGVPGEE551 pKa = 4.16MVAVGAAAVIGFLMLGNNN569 pKa = 3.91
MM1 pKa = 7.55GRR3 pKa = 11.84RR4 pKa = 11.84TFVKK8 pKa = 10.51GLGAAGAASVGLAHH22 pKa = 7.61DD23 pKa = 4.58RR24 pKa = 11.84GAARR28 pKa = 11.84NAEE31 pKa = 4.56AIAPLVGYY39 pKa = 10.29AIGAAAISAVGGIGVGWTLRR59 pKa = 11.84EE60 pKa = 4.03FEE62 pKa = 4.79VVGSDD67 pKa = 4.72DD68 pKa = 3.8PAEE71 pKa = 4.25GLTPDD76 pKa = 3.54VLRR79 pKa = 11.84NQLSDD84 pKa = 3.6SVVKK88 pKa = 10.61RR89 pKa = 11.84KK90 pKa = 10.36SNNQSTMVDD99 pKa = 3.25NQNILDD105 pKa = 4.25GVEE108 pKa = 3.87HH109 pKa = 6.22TAYY112 pKa = 9.83TEE114 pKa = 3.97AKK116 pKa = 8.8IAAIEE121 pKa = 3.95EE122 pKa = 4.43LNAGSSEE129 pKa = 4.28SAVLSAANSAIDD141 pKa = 3.52SYY143 pKa = 10.49EE144 pKa = 3.92TTVRR148 pKa = 11.84TNFYY152 pKa = 10.33KK153 pKa = 10.37SWNEE157 pKa = 3.67TVRR160 pKa = 11.84EE161 pKa = 4.25LEE163 pKa = 4.35AMTQTVIAHH172 pKa = 6.75ADD174 pKa = 3.22VGLSYY179 pKa = 9.8ITDD182 pKa = 3.94FGDD185 pKa = 3.42PRR187 pKa = 11.84FGNLASGTSPNTLKK201 pKa = 10.3DD202 pKa = 3.66TTVSMPDD209 pKa = 3.15GTNFTLLTFRR219 pKa = 11.84HH220 pKa = 4.74NTGWDD225 pKa = 3.43SGNAAYY231 pKa = 10.21SVVEE235 pKa = 4.01YY236 pKa = 10.77NPKK239 pKa = 10.38EE240 pKa = 4.22VVTSTNSNTYY250 pKa = 8.38NTVDD254 pKa = 2.97GTQYY258 pKa = 10.65MKK260 pKa = 10.67FSEE263 pKa = 4.31WNAVEE268 pKa = 4.33TEE270 pKa = 3.86MDD272 pKa = 3.59TVFQNVRR279 pKa = 11.84NGISTWVTNVYY290 pKa = 11.18GDD292 pKa = 3.84VQSGAIEE299 pKa = 4.05ISDD302 pKa = 3.5LVTPRR307 pKa = 11.84EE308 pKa = 4.24RR309 pKa = 11.84ATMMAQEE316 pKa = 4.65EE317 pKa = 4.92GMSQAIADD325 pKa = 5.12LIALNVPVDD334 pKa = 3.84AEE336 pKa = 4.23RR337 pKa = 11.84EE338 pKa = 4.21ATITIQDD345 pKa = 3.56TGATLPGTFALTDD358 pKa = 3.56SSDD361 pKa = 3.78GPLSAGQTYY370 pKa = 10.66DD371 pKa = 3.09PSTFSGDD378 pKa = 3.59VYY380 pKa = 9.75FTADD384 pKa = 3.16MSLVEE389 pKa = 4.75GPWDD393 pKa = 4.62AINSGVDD400 pKa = 2.88GGTITITSEE409 pKa = 4.15PYY411 pKa = 9.2EE412 pKa = 4.25GTAIEE417 pKa = 4.41VTTVEE422 pKa = 4.47SEE424 pKa = 4.64TVSVPAADD432 pKa = 3.35WTDD435 pKa = 3.4NGDD438 pKa = 3.85GTWSYY443 pKa = 11.33DD444 pKa = 3.16ASGDD448 pKa = 4.09LEE450 pKa = 4.42TTITNVDD457 pKa = 3.36SARR460 pKa = 11.84FVSTATEE467 pKa = 4.08TTYY470 pKa = 10.1DD471 pKa = 3.54TLQLKK476 pKa = 10.21GAFTVDD482 pKa = 2.98KK483 pKa = 10.64LVNKK487 pKa = 10.19QSGEE491 pKa = 4.3EE492 pKa = 4.01VSSTSFTSSEE502 pKa = 3.98PQTDD506 pKa = 3.34SNYY509 pKa = 8.7ITQDD513 pKa = 2.37EE514 pKa = 4.42WDD516 pKa = 3.49QLEE519 pKa = 4.23QQNKK523 pKa = 8.28EE524 pKa = 4.17LIEE527 pKa = 4.64KK528 pKa = 10.22YY529 pKa = 9.73EE530 pKa = 3.97QSQSGGGLDD539 pKa = 4.92LGGLDD544 pKa = 3.42MFGVPGEE551 pKa = 4.16MVAVGAAAVIGFLMLGNNN569 pKa = 3.91
Molecular weight: 60.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9ABP4|H9ABP4_9VIRU ORF3 OS=Halorubrum pleomorphic virus 6 OX=1156721 GN=ORF3 PE=4 SV=1
MM1 pKa = 6.31TTCPLTTHH9 pKa = 6.29YY10 pKa = 11.43ADD12 pKa = 3.46GRR14 pKa = 11.84RR15 pKa = 11.84PEE17 pKa = 4.63ADD19 pKa = 3.03ANRR22 pKa = 11.84SADD25 pKa = 3.67EE26 pKa = 4.04TRR28 pKa = 11.84SPGGEE33 pKa = 3.41RR34 pKa = 11.84AASQLLEE41 pKa = 3.98SAAAQEE47 pKa = 4.5MKK49 pKa = 10.57TPGTDD54 pKa = 3.02TVRR57 pKa = 11.84KK58 pKa = 8.29PGSPRR63 pKa = 11.84SLNPEE68 pKa = 3.49RR69 pKa = 11.84RR70 pKa = 11.84AKK72 pKa = 10.16RR73 pKa = 11.84AEE75 pKa = 3.82GNRR78 pKa = 11.84DD79 pKa = 2.6GSRR82 pKa = 11.84RR83 pKa = 11.84GRR85 pKa = 11.84QPGTASWPRR94 pKa = 11.84PEE96 pKa = 5.07SLDD99 pKa = 3.68LRR101 pKa = 11.84AAEE104 pKa = 4.63CSSCKK109 pKa = 9.18TKK111 pKa = 10.76RR112 pKa = 11.84SDD114 pKa = 2.8TRR116 pKa = 11.84ARR118 pKa = 11.84LEE120 pKa = 4.19AGATRR125 pKa = 11.84LL126 pKa = 3.69
MM1 pKa = 6.31TTCPLTTHH9 pKa = 6.29YY10 pKa = 11.43ADD12 pKa = 3.46GRR14 pKa = 11.84RR15 pKa = 11.84PEE17 pKa = 4.63ADD19 pKa = 3.03ANRR22 pKa = 11.84SADD25 pKa = 3.67EE26 pKa = 4.04TRR28 pKa = 11.84SPGGEE33 pKa = 3.41RR34 pKa = 11.84AASQLLEE41 pKa = 3.98SAAAQEE47 pKa = 4.5MKK49 pKa = 10.57TPGTDD54 pKa = 3.02TVRR57 pKa = 11.84KK58 pKa = 8.29PGSPRR63 pKa = 11.84SLNPEE68 pKa = 3.49RR69 pKa = 11.84RR70 pKa = 11.84AKK72 pKa = 10.16RR73 pKa = 11.84AEE75 pKa = 3.82GNRR78 pKa = 11.84DD79 pKa = 2.6GSRR82 pKa = 11.84RR83 pKa = 11.84GRR85 pKa = 11.84QPGTASWPRR94 pKa = 11.84PEE96 pKa = 5.07SLDD99 pKa = 3.68LRR101 pKa = 11.84AAEE104 pKa = 4.63CSSCKK109 pKa = 9.18TKK111 pKa = 10.76RR112 pKa = 11.84SDD114 pKa = 2.8TRR116 pKa = 11.84ARR118 pKa = 11.84LEE120 pKa = 4.19AGATRR125 pKa = 11.84LL126 pKa = 3.69
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2676 |
74 |
655 |
267.6 |
29.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.118 ± 0.686 | 0.71 ± 0.299 |
8.969 ± 1.466 | 6.577 ± 0.781 |
3.102 ± 0.45 | 8.034 ± 0.52 |
1.682 ± 0.508 | 3.7 ± 0.377 |
2.99 ± 0.584 | 8.819 ± 0.807 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.644 ± 0.217 | 3.363 ± 0.544 |
4.41 ± 0.47 | 2.99 ± 0.35 |
6.988 ± 1.334 | 7.549 ± 1.046 |
7.735 ± 0.963 | 7.362 ± 0.836 |
1.719 ± 0.175 | 2.541 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
