
Streptomyces phage SF1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
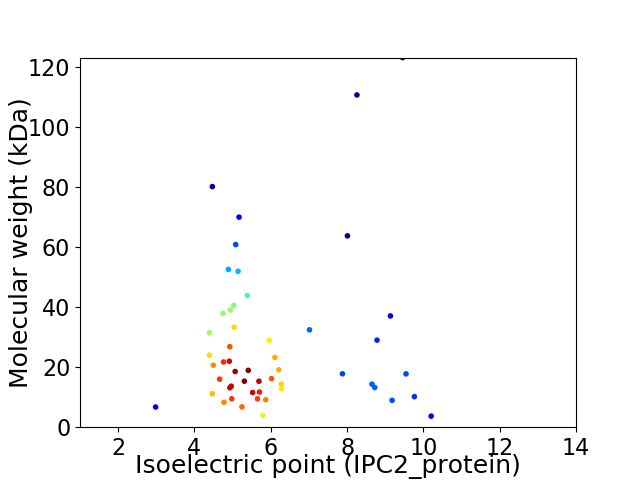
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
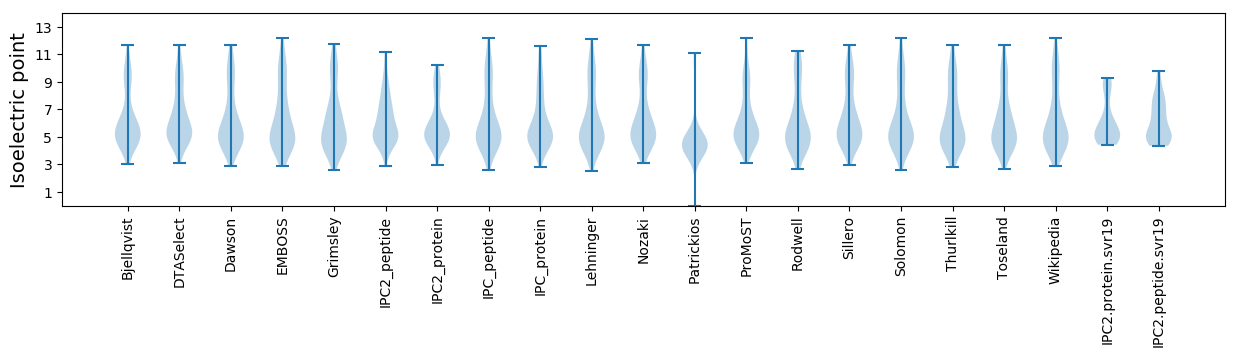
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1Y5S2|A0A0K1Y5S2_9CAUD Uncharacterized protein OS=Streptomyces phage SF1 OX=1690817 GN=SF1_210 PE=4 SV=1
MM1 pKa = 7.84AFPNPNLLPDD11 pKa = 4.53DD12 pKa = 4.66ASQFEE17 pKa = 4.85GATHH21 pKa = 6.26SWIDD25 pKa = 3.47PSNATLSIVSGQYY38 pKa = 8.58MAGSKK43 pKa = 8.77SLRR46 pKa = 11.84LTATAAGTASASTPYY61 pKa = 10.29VLSGLAPGKK70 pKa = 9.17TYY72 pKa = 10.44VARR75 pKa = 11.84IPIRR79 pKa = 11.84VSTASAGKK87 pKa = 7.58TATVRR92 pKa = 11.84MAFYY96 pKa = 10.62ADD98 pKa = 3.8SGPSIGTMDD107 pKa = 3.57AVVNLSATGTGWLPDD122 pKa = 3.61NFPTVSMVAPPAAAKK137 pKa = 10.29VRR139 pKa = 11.84MVVTVTGMAAGEE151 pKa = 4.19YY152 pKa = 10.51VNFDD156 pKa = 3.36DD157 pKa = 6.11CYY159 pKa = 10.9LGEE162 pKa = 4.84APLIPGNLYY171 pKa = 9.97PYY173 pKa = 8.69TVQSMEE179 pKa = 4.58SGVSTWGATGATTLWGDD196 pKa = 3.69GTRR199 pKa = 11.84SDD201 pKa = 3.75GFRR204 pKa = 11.84NLGLTATSAGVIAARR219 pKa = 11.84PNTYY223 pKa = 10.28VPVSAGAEE231 pKa = 4.13YY232 pKa = 10.75VGEE235 pKa = 4.0AWVYY239 pKa = 9.87SAVATTVDD247 pKa = 4.53ALIVWADD254 pKa = 3.43STGTEE259 pKa = 3.94VGRR262 pKa = 11.84TLVTRR267 pKa = 11.84DD268 pKa = 3.37ATSGAWSYY276 pKa = 10.49FIAVGVAPPNATQARR291 pKa = 11.84LFIQPKK297 pKa = 10.02ALATGDD303 pKa = 3.47TFYY306 pKa = 11.56VDD308 pKa = 3.51EE309 pKa = 5.1AALKK313 pKa = 9.11TSPNPTTNLLTYY325 pKa = 10.38DD326 pKa = 3.92EE327 pKa = 5.13YY328 pKa = 11.05STEE331 pKa = 4.08STLPPWTWDD340 pKa = 2.84GGVADD345 pKa = 4.74RR346 pKa = 11.84SYY348 pKa = 10.94FYY350 pKa = 10.7STITDD355 pKa = 2.75GRR357 pKa = 11.84YY358 pKa = 8.16IQRR361 pKa = 11.84VTPDD365 pKa = 3.07GPGYY369 pKa = 10.79LSGFMDD375 pKa = 3.76RR376 pKa = 11.84LVPVTEE382 pKa = 3.96GTTYY386 pKa = 10.45QLRR389 pKa = 11.84TLVLRR394 pKa = 11.84RR395 pKa = 11.84NPTAEE400 pKa = 4.85AISLTARR407 pKa = 11.84VRR409 pKa = 11.84IDD411 pKa = 2.87WYY413 pKa = 10.57DD414 pKa = 3.65DD415 pKa = 3.18NGTMVQADD423 pKa = 4.01VPDD426 pKa = 3.64QYY428 pKa = 11.77ATVTEE433 pKa = 4.38SVTYY437 pKa = 10.47SGVLLGEE444 pKa = 4.39TRR446 pKa = 11.84TCPPGATRR454 pKa = 11.84ARR456 pKa = 11.84LGVDD460 pKa = 2.69IDD462 pKa = 3.79HH463 pKa = 6.61TQTLAEE469 pKa = 4.77FYY471 pKa = 10.78YY472 pKa = 10.31VDD474 pKa = 3.41KK475 pKa = 10.99FQFYY479 pKa = 11.08VSDD482 pKa = 3.27SLYY485 pKa = 10.94AVTVSDD491 pKa = 3.63SRR493 pKa = 11.84GSIALQVNYY502 pKa = 10.2SPAGATSVSIRR513 pKa = 11.84RR514 pKa = 11.84MDD516 pKa = 3.64EE517 pKa = 4.53DD518 pKa = 4.08GSSTYY523 pKa = 10.2IRR525 pKa = 11.84GYY527 pKa = 9.17GQEE530 pKa = 4.16FNKK533 pKa = 10.39APYY536 pKa = 10.12SPGPIVAEE544 pKa = 4.71DD545 pKa = 3.72YY546 pKa = 7.97EE547 pKa = 5.6APLGTRR553 pKa = 11.84VWYY556 pKa = 10.17ALEE559 pKa = 3.94WFGASGSPVARR570 pKa = 11.84VFTQTVTGPVLEE582 pKa = 5.43DD583 pKa = 3.32GDD585 pKa = 4.53YY586 pKa = 11.52VWFKK590 pKa = 11.52SPGLPALNVQVMMEE604 pKa = 4.09TAIKK608 pKa = 9.47WEE610 pKa = 3.75RR611 pKa = 11.84AARR614 pKa = 11.84KK615 pKa = 7.64TSLTIVGRR623 pKa = 11.84KK624 pKa = 9.18NPVDD628 pKa = 3.18ITDD631 pKa = 3.31VRR633 pKa = 11.84GGRR636 pKa = 11.84TGSLSILVWDD646 pKa = 4.77EE647 pKa = 4.01SSHH650 pKa = 6.09VLFDD654 pKa = 3.57QLLDD658 pKa = 3.52SGLPVLIQAMPGYY671 pKa = 9.99GVAGNLFLSIGDD683 pKa = 3.53VSAEE687 pKa = 3.98NVLGVANEE695 pKa = 4.21PGWRR699 pKa = 11.84WALAVSEE706 pKa = 4.47IDD708 pKa = 3.84RR709 pKa = 11.84PDD711 pKa = 3.74GGLQGSAGRR720 pKa = 11.84TWQDD724 pKa = 2.98VMDD727 pKa = 5.39EE728 pKa = 4.23NADD731 pKa = 3.45WAAVDD736 pKa = 4.36AKK738 pKa = 9.91FATWAGVLTNNTT750 pKa = 3.56
MM1 pKa = 7.84AFPNPNLLPDD11 pKa = 4.53DD12 pKa = 4.66ASQFEE17 pKa = 4.85GATHH21 pKa = 6.26SWIDD25 pKa = 3.47PSNATLSIVSGQYY38 pKa = 8.58MAGSKK43 pKa = 8.77SLRR46 pKa = 11.84LTATAAGTASASTPYY61 pKa = 10.29VLSGLAPGKK70 pKa = 9.17TYY72 pKa = 10.44VARR75 pKa = 11.84IPIRR79 pKa = 11.84VSTASAGKK87 pKa = 7.58TATVRR92 pKa = 11.84MAFYY96 pKa = 10.62ADD98 pKa = 3.8SGPSIGTMDD107 pKa = 3.57AVVNLSATGTGWLPDD122 pKa = 3.61NFPTVSMVAPPAAAKK137 pKa = 10.29VRR139 pKa = 11.84MVVTVTGMAAGEE151 pKa = 4.19YY152 pKa = 10.51VNFDD156 pKa = 3.36DD157 pKa = 6.11CYY159 pKa = 10.9LGEE162 pKa = 4.84APLIPGNLYY171 pKa = 9.97PYY173 pKa = 8.69TVQSMEE179 pKa = 4.58SGVSTWGATGATTLWGDD196 pKa = 3.69GTRR199 pKa = 11.84SDD201 pKa = 3.75GFRR204 pKa = 11.84NLGLTATSAGVIAARR219 pKa = 11.84PNTYY223 pKa = 10.28VPVSAGAEE231 pKa = 4.13YY232 pKa = 10.75VGEE235 pKa = 4.0AWVYY239 pKa = 9.87SAVATTVDD247 pKa = 4.53ALIVWADD254 pKa = 3.43STGTEE259 pKa = 3.94VGRR262 pKa = 11.84TLVTRR267 pKa = 11.84DD268 pKa = 3.37ATSGAWSYY276 pKa = 10.49FIAVGVAPPNATQARR291 pKa = 11.84LFIQPKK297 pKa = 10.02ALATGDD303 pKa = 3.47TFYY306 pKa = 11.56VDD308 pKa = 3.51EE309 pKa = 5.1AALKK313 pKa = 9.11TSPNPTTNLLTYY325 pKa = 10.38DD326 pKa = 3.92EE327 pKa = 5.13YY328 pKa = 11.05STEE331 pKa = 4.08STLPPWTWDD340 pKa = 2.84GGVADD345 pKa = 4.74RR346 pKa = 11.84SYY348 pKa = 10.94FYY350 pKa = 10.7STITDD355 pKa = 2.75GRR357 pKa = 11.84YY358 pKa = 8.16IQRR361 pKa = 11.84VTPDD365 pKa = 3.07GPGYY369 pKa = 10.79LSGFMDD375 pKa = 3.76RR376 pKa = 11.84LVPVTEE382 pKa = 3.96GTTYY386 pKa = 10.45QLRR389 pKa = 11.84TLVLRR394 pKa = 11.84RR395 pKa = 11.84NPTAEE400 pKa = 4.85AISLTARR407 pKa = 11.84VRR409 pKa = 11.84IDD411 pKa = 2.87WYY413 pKa = 10.57DD414 pKa = 3.65DD415 pKa = 3.18NGTMVQADD423 pKa = 4.01VPDD426 pKa = 3.64QYY428 pKa = 11.77ATVTEE433 pKa = 4.38SVTYY437 pKa = 10.47SGVLLGEE444 pKa = 4.39TRR446 pKa = 11.84TCPPGATRR454 pKa = 11.84ARR456 pKa = 11.84LGVDD460 pKa = 2.69IDD462 pKa = 3.79HH463 pKa = 6.61TQTLAEE469 pKa = 4.77FYY471 pKa = 10.78YY472 pKa = 10.31VDD474 pKa = 3.41KK475 pKa = 10.99FQFYY479 pKa = 11.08VSDD482 pKa = 3.27SLYY485 pKa = 10.94AVTVSDD491 pKa = 3.63SRR493 pKa = 11.84GSIALQVNYY502 pKa = 10.2SPAGATSVSIRR513 pKa = 11.84RR514 pKa = 11.84MDD516 pKa = 3.64EE517 pKa = 4.53DD518 pKa = 4.08GSSTYY523 pKa = 10.2IRR525 pKa = 11.84GYY527 pKa = 9.17GQEE530 pKa = 4.16FNKK533 pKa = 10.39APYY536 pKa = 10.12SPGPIVAEE544 pKa = 4.71DD545 pKa = 3.72YY546 pKa = 7.97EE547 pKa = 5.6APLGTRR553 pKa = 11.84VWYY556 pKa = 10.17ALEE559 pKa = 3.94WFGASGSPVARR570 pKa = 11.84VFTQTVTGPVLEE582 pKa = 5.43DD583 pKa = 3.32GDD585 pKa = 4.53YY586 pKa = 11.52VWFKK590 pKa = 11.52SPGLPALNVQVMMEE604 pKa = 4.09TAIKK608 pKa = 9.47WEE610 pKa = 3.75RR611 pKa = 11.84AARR614 pKa = 11.84KK615 pKa = 7.64TSLTIVGRR623 pKa = 11.84KK624 pKa = 9.18NPVDD628 pKa = 3.18ITDD631 pKa = 3.31VRR633 pKa = 11.84GGRR636 pKa = 11.84TGSLSILVWDD646 pKa = 4.77EE647 pKa = 4.01SSHH650 pKa = 6.09VLFDD654 pKa = 3.57QLLDD658 pKa = 3.52SGLPVLIQAMPGYY671 pKa = 9.99GVAGNLFLSIGDD683 pKa = 3.53VSAEE687 pKa = 3.98NVLGVANEE695 pKa = 4.21PGWRR699 pKa = 11.84WALAVSEE706 pKa = 4.47IDD708 pKa = 3.84RR709 pKa = 11.84PDD711 pKa = 3.74GGLQGSAGRR720 pKa = 11.84TWQDD724 pKa = 2.98VMDD727 pKa = 5.39EE728 pKa = 4.23NADD731 pKa = 3.45WAAVDD736 pKa = 4.36AKK738 pKa = 9.91FATWAGVLTNNTT750 pKa = 3.56
Molecular weight: 80.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1Y5T7|A0A0K1Y5T7_9CAUD Uncharacterized protein OS=Streptomyces phage SF1 OX=1690817 GN=SF1_360 PE=4 SV=1
MM1 pKa = 7.11TMARR5 pKa = 11.84PSFRR9 pKa = 11.84PNRR12 pKa = 11.84RR13 pKa = 11.84NIAGFLKK20 pKa = 10.16TPATHH25 pKa = 7.25ALIEE29 pKa = 4.43RR30 pKa = 11.84KK31 pKa = 7.64TRR33 pKa = 11.84AAEE36 pKa = 4.14SAASAAARR44 pKa = 11.84SDD46 pKa = 2.9GWGGQFRR53 pKa = 11.84TDD55 pKa = 3.23VEE57 pKa = 4.57TGDD60 pKa = 3.2KK61 pKa = 9.89RR62 pKa = 11.84ARR64 pKa = 11.84GAVIGDD70 pKa = 3.53YY71 pKa = 8.43STPYY75 pKa = 9.98PEE77 pKa = 4.13VSRR80 pKa = 11.84RR81 pKa = 11.84ALLRR85 pKa = 11.84ALDD88 pKa = 3.81AARR91 pKa = 11.84GAEE94 pKa = 3.93
MM1 pKa = 7.11TMARR5 pKa = 11.84PSFRR9 pKa = 11.84PNRR12 pKa = 11.84RR13 pKa = 11.84NIAGFLKK20 pKa = 10.16TPATHH25 pKa = 7.25ALIEE29 pKa = 4.43RR30 pKa = 11.84KK31 pKa = 7.64TRR33 pKa = 11.84AAEE36 pKa = 4.14SAASAAARR44 pKa = 11.84SDD46 pKa = 2.9GWGGQFRR53 pKa = 11.84TDD55 pKa = 3.23VEE57 pKa = 4.57TGDD60 pKa = 3.2KK61 pKa = 9.89RR62 pKa = 11.84ARR64 pKa = 11.84GAVIGDD70 pKa = 3.53YY71 pKa = 8.43STPYY75 pKa = 9.98PEE77 pKa = 4.13VSRR80 pKa = 11.84RR81 pKa = 11.84ALLRR85 pKa = 11.84ALDD88 pKa = 3.81AARR91 pKa = 11.84GAEE94 pKa = 3.93
Molecular weight: 10.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13540 |
32 |
1190 |
260.4 |
27.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.888 ± 0.447 | 0.598 ± 0.104 |
6.551 ± 0.302 | 5.244 ± 0.422 |
2.386 ± 0.187 | 10.074 ± 0.513 |
1.396 ± 0.2 | 3.538 ± 0.143 |
3.405 ± 0.383 | 8.235 ± 0.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.92 ± 0.136 | 2.563 ± 0.191 |
5.643 ± 0.325 | 3.124 ± 0.161 |
7.186 ± 0.433 | 5.465 ± 0.247 |
6.78 ± 0.415 | 8.227 ± 0.282 |
2.157 ± 0.149 | 2.622 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
