
Bluetongue virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; Bluetongue virus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
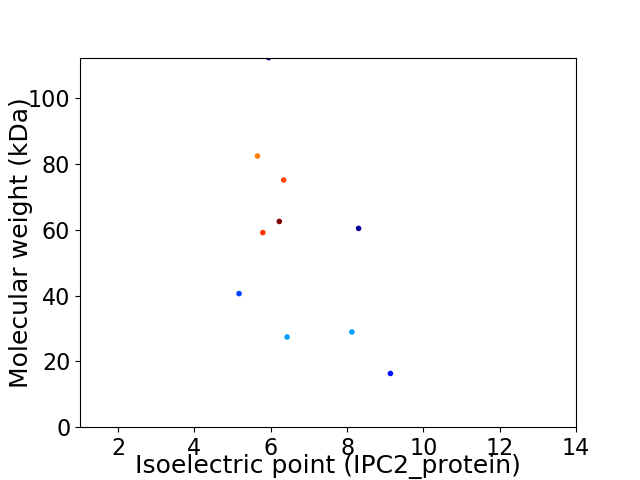
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
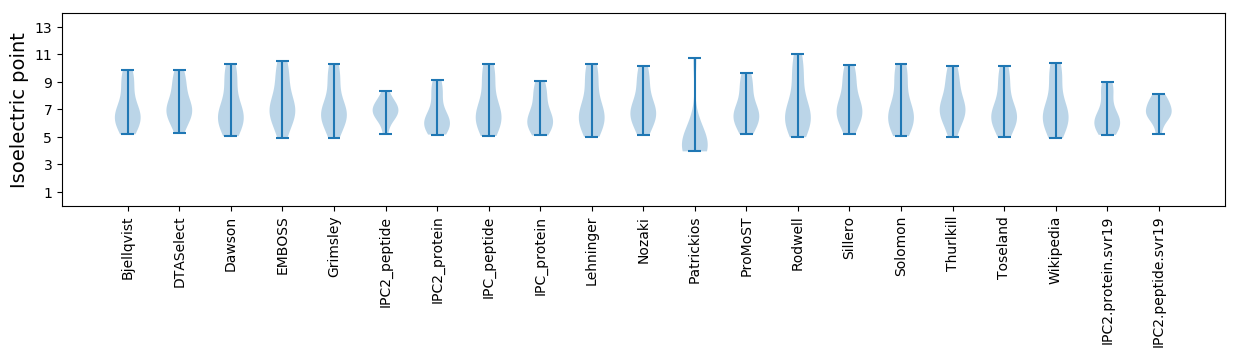
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V0CK01|A0A1V0CK01_9REOV Outer capsid protein VP5 OS=Bluetongue virus 3 OX=36423 PE=3 SV=1
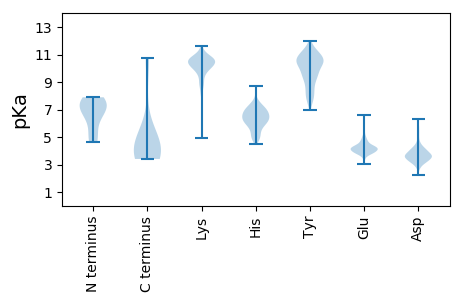
MM1 pKa = 7.04EE2 pKa = 4.1QKK4 pKa = 9.85QRR6 pKa = 11.84RR7 pKa = 11.84FTKK10 pKa = 10.67NIFVLDD16 pKa = 4.23ANAKK20 pKa = 4.94TLCGAIAKK28 pKa = 9.87LSSLPYY34 pKa = 9.45CQIKK38 pKa = 9.33IGRR41 pKa = 11.84VIAFKK46 pKa = 10.12PVKK49 pKa = 10.12NPEE52 pKa = 3.87PKK54 pKa = 10.38GYY56 pKa = 10.08VLNVPGPGAYY66 pKa = 9.6RR67 pKa = 11.84IQDD70 pKa = 3.57GQDD73 pKa = 3.18IISLMLTPHH82 pKa = 5.88GVEE85 pKa = 3.73ATTEE89 pKa = 3.8RR90 pKa = 11.84WEE92 pKa = 3.82EE93 pKa = 3.85WKK95 pKa = 11.01FEE97 pKa = 4.21GVSVTPMATRR107 pKa = 11.84VQHH110 pKa = 5.97NGVMVDD116 pKa = 5.38AEE118 pKa = 4.1IKK120 pKa = 8.19YY121 pKa = 9.36CKK123 pKa = 10.33GMGIVQPYY131 pKa = 8.14MRR133 pKa = 11.84NDD135 pKa = 3.34FDD137 pKa = 5.32RR138 pKa = 11.84NEE140 pKa = 4.16MPDD143 pKa = 3.43LPGVMRR149 pKa = 11.84SNYY152 pKa = 9.42DD153 pKa = 3.28VRR155 pKa = 11.84EE156 pKa = 3.94LRR158 pKa = 11.84QKK160 pKa = 10.23IKK162 pKa = 10.85NEE164 pKa = 3.89RR165 pKa = 11.84EE166 pKa = 3.82SAPRR170 pKa = 11.84LQVQSVAPRR179 pKa = 11.84EE180 pKa = 4.1EE181 pKa = 4.34SRR183 pKa = 11.84WMDD186 pKa = 3.74DD187 pKa = 4.45DD188 pKa = 3.79EE189 pKa = 6.59AKK191 pKa = 10.56VDD193 pKa = 3.91EE194 pKa = 4.88EE195 pKa = 4.35AKK197 pKa = 10.84EE198 pKa = 4.14MIPGTSRR205 pKa = 11.84LEE207 pKa = 3.96KK208 pKa = 10.49LRR210 pKa = 11.84EE211 pKa = 3.81ARR213 pKa = 11.84SNVFKK218 pKa = 10.71EE219 pKa = 4.2VEE221 pKa = 4.44AGINWNLDD229 pKa = 3.37EE230 pKa = 5.9KK231 pKa = 11.28DD232 pKa = 3.5EE233 pKa = 4.18EE234 pKa = 4.5DD235 pKa = 3.59RR236 pKa = 11.84DD237 pKa = 3.75EE238 pKa = 6.46RR239 pKa = 11.84EE240 pKa = 4.63DD241 pKa = 3.64EE242 pKa = 4.3EE243 pKa = 4.37QAKK246 pKa = 8.35TLSDD250 pKa = 3.97DD251 pKa = 4.35DD252 pKa = 4.22EE253 pKa = 4.93QGEE256 pKa = 4.29DD257 pKa = 3.59TSDD260 pKa = 3.7DD261 pKa = 3.9EE262 pKa = 5.02HH263 pKa = 6.98PKK265 pKa = 8.37THH267 pKa = 5.79ITKK270 pKa = 10.22EE271 pKa = 4.16YY272 pKa = 8.56IEE274 pKa = 4.81KK275 pKa = 10.13VAKK278 pKa = 9.73QIKK281 pKa = 10.13LKK283 pKa = 10.47DD284 pKa = 3.66EE285 pKa = 4.41RR286 pKa = 11.84FMSLSSAMPQASGGFDD302 pKa = 3.1RR303 pKa = 11.84MIVTKK308 pKa = 10.26KK309 pKa = 10.67LKK311 pKa = 8.0WQNVPLYY318 pKa = 10.72CFDD321 pKa = 4.17EE322 pKa = 4.26SSKK325 pKa = 10.62RR326 pKa = 11.84YY327 pKa = 8.95EE328 pKa = 4.11LQCVGACEE336 pKa = 3.83RR337 pKa = 11.84VAFVSKK343 pKa = 10.81DD344 pKa = 3.14MSLIILPVGVV354 pKa = 3.86
MM1 pKa = 7.04EE2 pKa = 4.1QKK4 pKa = 9.85QRR6 pKa = 11.84RR7 pKa = 11.84FTKK10 pKa = 10.67NIFVLDD16 pKa = 4.23ANAKK20 pKa = 4.94TLCGAIAKK28 pKa = 9.87LSSLPYY34 pKa = 9.45CQIKK38 pKa = 9.33IGRR41 pKa = 11.84VIAFKK46 pKa = 10.12PVKK49 pKa = 10.12NPEE52 pKa = 3.87PKK54 pKa = 10.38GYY56 pKa = 10.08VLNVPGPGAYY66 pKa = 9.6RR67 pKa = 11.84IQDD70 pKa = 3.57GQDD73 pKa = 3.18IISLMLTPHH82 pKa = 5.88GVEE85 pKa = 3.73ATTEE89 pKa = 3.8RR90 pKa = 11.84WEE92 pKa = 3.82EE93 pKa = 3.85WKK95 pKa = 11.01FEE97 pKa = 4.21GVSVTPMATRR107 pKa = 11.84VQHH110 pKa = 5.97NGVMVDD116 pKa = 5.38AEE118 pKa = 4.1IKK120 pKa = 8.19YY121 pKa = 9.36CKK123 pKa = 10.33GMGIVQPYY131 pKa = 8.14MRR133 pKa = 11.84NDD135 pKa = 3.34FDD137 pKa = 5.32RR138 pKa = 11.84NEE140 pKa = 4.16MPDD143 pKa = 3.43LPGVMRR149 pKa = 11.84SNYY152 pKa = 9.42DD153 pKa = 3.28VRR155 pKa = 11.84EE156 pKa = 3.94LRR158 pKa = 11.84QKK160 pKa = 10.23IKK162 pKa = 10.85NEE164 pKa = 3.89RR165 pKa = 11.84EE166 pKa = 3.82SAPRR170 pKa = 11.84LQVQSVAPRR179 pKa = 11.84EE180 pKa = 4.1EE181 pKa = 4.34SRR183 pKa = 11.84WMDD186 pKa = 3.74DD187 pKa = 4.45DD188 pKa = 3.79EE189 pKa = 6.59AKK191 pKa = 10.56VDD193 pKa = 3.91EE194 pKa = 4.88EE195 pKa = 4.35AKK197 pKa = 10.84EE198 pKa = 4.14MIPGTSRR205 pKa = 11.84LEE207 pKa = 3.96KK208 pKa = 10.49LRR210 pKa = 11.84EE211 pKa = 3.81ARR213 pKa = 11.84SNVFKK218 pKa = 10.71EE219 pKa = 4.2VEE221 pKa = 4.44AGINWNLDD229 pKa = 3.37EE230 pKa = 5.9KK231 pKa = 11.28DD232 pKa = 3.5EE233 pKa = 4.18EE234 pKa = 4.5DD235 pKa = 3.59RR236 pKa = 11.84DD237 pKa = 3.75EE238 pKa = 6.46RR239 pKa = 11.84EE240 pKa = 4.63DD241 pKa = 3.64EE242 pKa = 4.3EE243 pKa = 4.37QAKK246 pKa = 8.35TLSDD250 pKa = 3.97DD251 pKa = 4.35DD252 pKa = 4.22EE253 pKa = 4.93QGEE256 pKa = 4.29DD257 pKa = 3.59TSDD260 pKa = 3.7DD261 pKa = 3.9EE262 pKa = 5.02HH263 pKa = 6.98PKK265 pKa = 8.37THH267 pKa = 5.79ITKK270 pKa = 10.22EE271 pKa = 4.16YY272 pKa = 8.56IEE274 pKa = 4.81KK275 pKa = 10.13VAKK278 pKa = 9.73QIKK281 pKa = 10.13LKK283 pKa = 10.47DD284 pKa = 3.66EE285 pKa = 4.41RR286 pKa = 11.84FMSLSSAMPQASGGFDD302 pKa = 3.1RR303 pKa = 11.84MIVTKK308 pKa = 10.26KK309 pKa = 10.67LKK311 pKa = 8.0WQNVPLYY318 pKa = 10.72CFDD321 pKa = 4.17EE322 pKa = 4.26SSKK325 pKa = 10.62RR326 pKa = 11.84YY327 pKa = 8.95EE328 pKa = 4.11LQCVGACEE336 pKa = 3.83RR337 pKa = 11.84VAFVSKK343 pKa = 10.81DD344 pKa = 3.14MSLIILPVGVV354 pKa = 3.86
Molecular weight: 40.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0CK00|A0A1V0CK00_9REOV Non-structural protein NS2 OS=Bluetongue virus 3 OX=36423 PE=4 SV=1
QQ1 pKa = 5.09TQKK4 pKa = 11.21AEE6 pKa = 4.0KK7 pKa = 10.19AAFASYY13 pKa = 11.37AEE15 pKa = 4.24AFRR18 pKa = 11.84DD19 pKa = 4.13DD20 pKa = 3.2VRR22 pKa = 11.84LRR24 pKa = 11.84QIKK27 pKa = 9.78RR28 pKa = 11.84HH29 pKa = 4.98VNEE32 pKa = 4.23QILPKK37 pKa = 10.44LKK39 pKa = 10.51SDD41 pKa = 4.47LGGLKK46 pKa = 9.95KK47 pKa = 10.16KK48 pKa = 10.37RR49 pKa = 11.84AIIHH53 pKa = 4.9MTLLIAAVVALLTSVCTLSSDD74 pKa = 3.16MSVAFKK80 pKa = 11.33LNGTSAEE87 pKa = 3.85IPQWFKK93 pKa = 11.52SLNPMLGVVNLGATFLMMVCAKK115 pKa = 9.83SEE117 pKa = 3.92RR118 pKa = 11.84SLNQQIDD125 pKa = 4.05MIKK128 pKa = 10.21KK129 pKa = 9.45EE130 pKa = 4.14VMKK133 pKa = 10.61KK134 pKa = 8.85QSYY137 pKa = 9.19NDD139 pKa = 3.56AVRR142 pKa = 11.84MSFTT146 pKa = 3.63
QQ1 pKa = 5.09TQKK4 pKa = 11.21AEE6 pKa = 4.0KK7 pKa = 10.19AAFASYY13 pKa = 11.37AEE15 pKa = 4.24AFRR18 pKa = 11.84DD19 pKa = 4.13DD20 pKa = 3.2VRR22 pKa = 11.84LRR24 pKa = 11.84QIKK27 pKa = 9.78RR28 pKa = 11.84HH29 pKa = 4.98VNEE32 pKa = 4.23QILPKK37 pKa = 10.44LKK39 pKa = 10.51SDD41 pKa = 4.47LGGLKK46 pKa = 9.95KK47 pKa = 10.16KK48 pKa = 10.37RR49 pKa = 11.84AIIHH53 pKa = 4.9MTLLIAAVVALLTSVCTLSSDD74 pKa = 3.16MSVAFKK80 pKa = 11.33LNGTSAEE87 pKa = 3.85IPQWFKK93 pKa = 11.52SLNPMLGVVNLGATFLMMVCAKK115 pKa = 9.83SEE117 pKa = 3.92RR118 pKa = 11.84SLNQQIDD125 pKa = 4.05MIKK128 pKa = 10.21KK129 pKa = 9.45EE130 pKa = 4.14VMKK133 pKa = 10.61KK134 pKa = 8.85QSYY137 pKa = 9.19NDD139 pKa = 3.56AVRR142 pKa = 11.84MSFTT146 pKa = 3.63
Molecular weight: 16.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4930 |
146 |
959 |
493.0 |
56.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.856 ± 0.623 | 1.217 ± 0.294 |
6.308 ± 0.363 | 7.241 ± 0.657 |
3.834 ± 0.237 | 5.497 ± 0.596 |
2.394 ± 0.23 | 6.633 ± 0.285 |
6.065 ± 0.808 | 8.479 ± 0.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.671 ± 0.329 | 3.753 ± 0.328 |
4.178 ± 0.322 | 3.773 ± 0.349 |
7.059 ± 0.309 | 5.74 ± 0.324 |
4.97 ± 0.382 | 7.039 ± 0.204 |
1.46 ± 0.214 | 3.834 ± 0.296 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
