
Salipaludibacillus aurantiacus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Salipaludibacillus
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
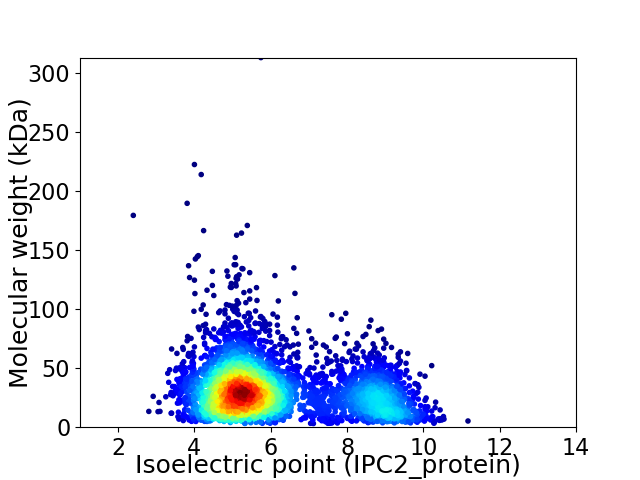
Virtual 2D-PAGE plot for 4227 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9TBN7|A0A1H9TBN7_9BACI Uncharacterized protein OS=Salipaludibacillus aurantiacus OX=1601833 GN=SAMN05518684_105206 PE=4 SV=1
MM1 pKa = 7.21KK2 pKa = 10.28KK3 pKa = 10.23KK4 pKa = 8.2YY5 pKa = 9.16TSMFALLLTGSLILAACGEE24 pKa = 4.33GDD26 pKa = 5.07DD27 pKa = 5.6NGNDD31 pKa = 3.56GNNAGGNNNAEE42 pKa = 4.06EE43 pKa = 4.52NEE45 pKa = 4.04ANDD48 pKa = 4.65NGGGDD53 pKa = 4.28DD54 pKa = 4.27NAAADD59 pKa = 3.95TDD61 pKa = 3.98VGNLQLGTGSTGGTYY76 pKa = 10.26YY77 pKa = 10.54PLGQEE82 pKa = 3.91MATVINNNIEE92 pKa = 3.77VDD94 pKa = 3.83GFDD97 pKa = 4.05LSAVSSGASVDD108 pKa = 3.82NIISIFQGEE117 pKa = 4.5MDD119 pKa = 4.37LGMSVHH125 pKa = 6.72LPALDD130 pKa = 3.74SLTGDD135 pKa = 3.53GDD137 pKa = 4.1FEE139 pKa = 4.43GVEE142 pKa = 3.88VDD144 pKa = 3.19NFGFMGHH151 pKa = 7.23IYY153 pKa = 9.81PEE155 pKa = 4.03VMQIVTGADD164 pKa = 3.09SGIEE168 pKa = 4.16SIGDD172 pKa = 3.66LKK174 pKa = 11.21DD175 pKa = 3.29KK176 pKa = 10.78SVAIGPPGSGTQAAAKK192 pKa = 9.84LVLEE196 pKa = 4.77AYY198 pKa = 10.07GLEE201 pKa = 4.52DD202 pKa = 3.69GDD204 pKa = 3.65YY205 pKa = 11.03DD206 pKa = 4.22AYY208 pKa = 10.73EE209 pKa = 4.29EE210 pKa = 4.81GFGDD214 pKa = 3.58AAGRR218 pKa = 11.84IQDD221 pKa = 3.99GNLDD225 pKa = 3.52ASFGLLGLPAGSIEE239 pKa = 4.13EE240 pKa = 4.16LSISRR245 pKa = 11.84DD246 pKa = 2.97IRR248 pKa = 11.84LLPISDD254 pKa = 4.12EE255 pKa = 4.05GLDD258 pKa = 3.81YY259 pKa = 11.05IEE261 pKa = 5.04EE262 pKa = 4.17NSGYY266 pKa = 10.08EE267 pKa = 4.11ALNIPADD274 pKa = 3.76SYY276 pKa = 12.01DD277 pKa = 4.52FMDD280 pKa = 4.76EE281 pKa = 3.92DD282 pKa = 4.04VQAITAYY289 pKa = 10.16AILVGSTDD297 pKa = 3.67TIDD300 pKa = 3.63EE301 pKa = 4.16EE302 pKa = 4.47LGYY305 pKa = 10.46EE306 pKa = 3.68ITKK309 pKa = 10.82ALFEE313 pKa = 4.37HH314 pKa = 6.63SGDD317 pKa = 3.31ISHH320 pKa = 7.27PQGDD324 pKa = 4.27HH325 pKa = 5.11LTKK328 pKa = 10.74EE329 pKa = 3.95NALNGSNGLPIHH341 pKa = 6.9PGAEE345 pKa = 3.52RR346 pKa = 11.84YY347 pKa = 9.37FEE349 pKa = 4.27EE350 pKa = 4.92EE351 pKa = 4.99GITEE355 pKa = 4.02
MM1 pKa = 7.21KK2 pKa = 10.28KK3 pKa = 10.23KK4 pKa = 8.2YY5 pKa = 9.16TSMFALLLTGSLILAACGEE24 pKa = 4.33GDD26 pKa = 5.07DD27 pKa = 5.6NGNDD31 pKa = 3.56GNNAGGNNNAEE42 pKa = 4.06EE43 pKa = 4.52NEE45 pKa = 4.04ANDD48 pKa = 4.65NGGGDD53 pKa = 4.28DD54 pKa = 4.27NAAADD59 pKa = 3.95TDD61 pKa = 3.98VGNLQLGTGSTGGTYY76 pKa = 10.26YY77 pKa = 10.54PLGQEE82 pKa = 3.91MATVINNNIEE92 pKa = 3.77VDD94 pKa = 3.83GFDD97 pKa = 4.05LSAVSSGASVDD108 pKa = 3.82NIISIFQGEE117 pKa = 4.5MDD119 pKa = 4.37LGMSVHH125 pKa = 6.72LPALDD130 pKa = 3.74SLTGDD135 pKa = 3.53GDD137 pKa = 4.1FEE139 pKa = 4.43GVEE142 pKa = 3.88VDD144 pKa = 3.19NFGFMGHH151 pKa = 7.23IYY153 pKa = 9.81PEE155 pKa = 4.03VMQIVTGADD164 pKa = 3.09SGIEE168 pKa = 4.16SIGDD172 pKa = 3.66LKK174 pKa = 11.21DD175 pKa = 3.29KK176 pKa = 10.78SVAIGPPGSGTQAAAKK192 pKa = 9.84LVLEE196 pKa = 4.77AYY198 pKa = 10.07GLEE201 pKa = 4.52DD202 pKa = 3.69GDD204 pKa = 3.65YY205 pKa = 11.03DD206 pKa = 4.22AYY208 pKa = 10.73EE209 pKa = 4.29EE210 pKa = 4.81GFGDD214 pKa = 3.58AAGRR218 pKa = 11.84IQDD221 pKa = 3.99GNLDD225 pKa = 3.52ASFGLLGLPAGSIEE239 pKa = 4.13EE240 pKa = 4.16LSISRR245 pKa = 11.84DD246 pKa = 2.97IRR248 pKa = 11.84LLPISDD254 pKa = 4.12EE255 pKa = 4.05GLDD258 pKa = 3.81YY259 pKa = 11.05IEE261 pKa = 5.04EE262 pKa = 4.17NSGYY266 pKa = 10.08EE267 pKa = 4.11ALNIPADD274 pKa = 3.76SYY276 pKa = 12.01DD277 pKa = 4.52FMDD280 pKa = 4.76EE281 pKa = 3.92DD282 pKa = 4.04VQAITAYY289 pKa = 10.16AILVGSTDD297 pKa = 3.67TIDD300 pKa = 3.63EE301 pKa = 4.16EE302 pKa = 4.47LGYY305 pKa = 10.46EE306 pKa = 3.68ITKK309 pKa = 10.82ALFEE313 pKa = 4.37HH314 pKa = 6.63SGDD317 pKa = 3.31ISHH320 pKa = 7.27PQGDD324 pKa = 4.27HH325 pKa = 5.11LTKK328 pKa = 10.74EE329 pKa = 3.95NALNGSNGLPIHH341 pKa = 6.9PGAEE345 pKa = 3.52RR346 pKa = 11.84YY347 pKa = 9.37FEE349 pKa = 4.27EE350 pKa = 4.92EE351 pKa = 4.99GITEE355 pKa = 4.02
Molecular weight: 37.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9VN47|A0A1H9VN47_9BACI Putative sporulation protein YyaC OS=Salipaludibacillus aurantiacus OX=1601833 GN=SAMN05518684_111103 PE=4 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.34KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.17VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.34KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.17VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1241661 |
29 |
2742 |
293.7 |
32.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.282 ± 0.052 | 0.669 ± 0.011 |
5.528 ± 0.06 | 8.189 ± 0.053 |
4.518 ± 0.03 | 7.197 ± 0.043 |
2.173 ± 0.019 | 6.984 ± 0.036 |
6.388 ± 0.045 | 9.56 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.855 ± 0.019 | 4.225 ± 0.027 |
3.803 ± 0.021 | 3.497 ± 0.025 |
4.284 ± 0.029 | 5.984 ± 0.029 |
5.358 ± 0.024 | 7.016 ± 0.032 |
1.062 ± 0.016 | 3.427 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
