
Faeces associated gemycircularvirus 22
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus bovas1; Bovine associated gemycircularvirus 1
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
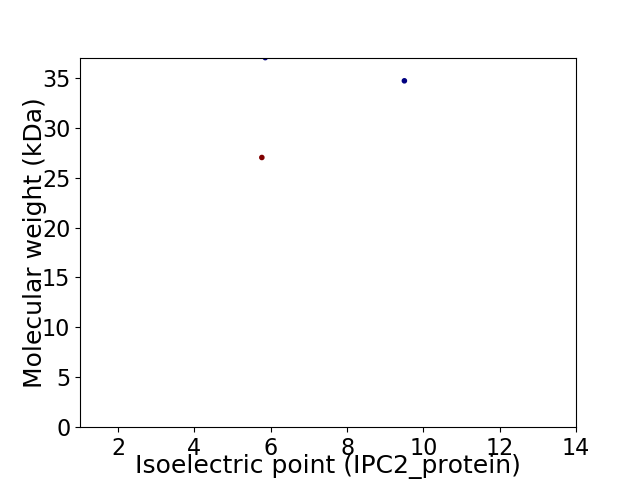
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
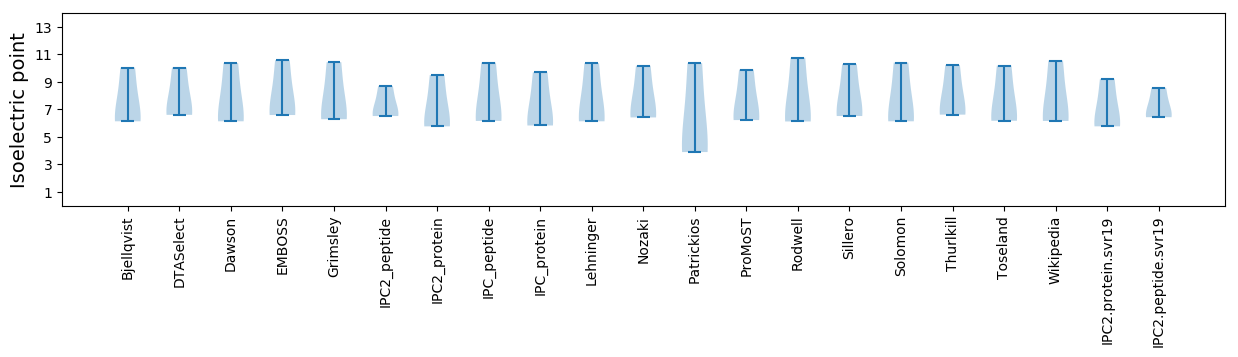
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161J5P3|A0A161J5P3_9VIRU RepA OS=Faeces associated gemycircularvirus 22 OX=1843742 PE=3 SV=1
MM1 pKa = 7.81PFHH4 pKa = 6.86FGARR8 pKa = 11.84YY9 pKa = 9.91ALITYY14 pKa = 7.36AQCNALDD21 pKa = 3.87GFRR24 pKa = 11.84VMDD27 pKa = 4.02HH28 pKa = 6.63FSGLGAEE35 pKa = 4.62CIVGRR40 pKa = 11.84EE41 pKa = 3.86VHH43 pKa = 6.72ADD45 pKa = 3.32GGIHH49 pKa = 6.0LHH51 pKa = 6.36CFIDD55 pKa = 4.11FGRR58 pKa = 11.84KK59 pKa = 7.81FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84TDD66 pKa = 2.64IFDD69 pKa = 3.14VDD71 pKa = 3.53GRR73 pKa = 11.84HH74 pKa = 6.16PNIAPSYY81 pKa = 6.47GTPWRR86 pKa = 11.84GYY88 pKa = 10.77DD89 pKa = 3.55YY90 pKa = 10.79AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.29VICGGLEE103 pKa = 3.73RR104 pKa = 11.84PEE106 pKa = 4.01EE107 pKa = 4.12PRR109 pKa = 11.84SKK111 pKa = 10.46RR112 pKa = 11.84VTKK115 pKa = 10.54DD116 pKa = 2.49WDD118 pKa = 3.16PWTEE122 pKa = 3.55ITNARR127 pKa = 11.84DD128 pKa = 3.58RR129 pKa = 11.84EE130 pKa = 4.49HH131 pKa = 6.41FWEE134 pKa = 4.87LVHH137 pKa = 8.03HH138 pKa = 7.16LDD140 pKa = 4.88PKK142 pKa = 10.88AAACNYY148 pKa = 8.82GQLAKK153 pKa = 10.78YY154 pKa = 10.48ADD156 pKa = 3.32WRR158 pKa = 11.84FAAKK162 pKa = 9.95PPVYY166 pKa = 9.39EE167 pKa = 4.2SPGGIEE173 pKa = 4.29FVGGDD178 pKa = 3.31VDD180 pKa = 6.46GRR182 pKa = 11.84DD183 pKa = 3.34AWCDD187 pKa = 3.17QSGIRR192 pKa = 11.84SGDD195 pKa = 3.6PLIGTSVRR203 pKa = 11.84SPRR206 pKa = 11.84LPSSGTIGGVCLGLAPQGGPYY227 pKa = 9.23TPPDD231 pKa = 3.56GAEE234 pKa = 4.08TSLSRR239 pKa = 11.84CNANCRR245 pKa = 3.61
MM1 pKa = 7.81PFHH4 pKa = 6.86FGARR8 pKa = 11.84YY9 pKa = 9.91ALITYY14 pKa = 7.36AQCNALDD21 pKa = 3.87GFRR24 pKa = 11.84VMDD27 pKa = 4.02HH28 pKa = 6.63FSGLGAEE35 pKa = 4.62CIVGRR40 pKa = 11.84EE41 pKa = 3.86VHH43 pKa = 6.72ADD45 pKa = 3.32GGIHH49 pKa = 6.0LHH51 pKa = 6.36CFIDD55 pKa = 4.11FGRR58 pKa = 11.84KK59 pKa = 7.81FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84TDD66 pKa = 2.64IFDD69 pKa = 3.14VDD71 pKa = 3.53GRR73 pKa = 11.84HH74 pKa = 6.16PNIAPSYY81 pKa = 6.47GTPWRR86 pKa = 11.84GYY88 pKa = 10.77DD89 pKa = 3.55YY90 pKa = 10.79AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.29VICGGLEE103 pKa = 3.73RR104 pKa = 11.84PEE106 pKa = 4.01EE107 pKa = 4.12PRR109 pKa = 11.84SKK111 pKa = 10.46RR112 pKa = 11.84VTKK115 pKa = 10.54DD116 pKa = 2.49WDD118 pKa = 3.16PWTEE122 pKa = 3.55ITNARR127 pKa = 11.84DD128 pKa = 3.58RR129 pKa = 11.84EE130 pKa = 4.49HH131 pKa = 6.41FWEE134 pKa = 4.87LVHH137 pKa = 8.03HH138 pKa = 7.16LDD140 pKa = 4.88PKK142 pKa = 10.88AAACNYY148 pKa = 8.82GQLAKK153 pKa = 10.78YY154 pKa = 10.48ADD156 pKa = 3.32WRR158 pKa = 11.84FAAKK162 pKa = 9.95PPVYY166 pKa = 9.39EE167 pKa = 4.2SPGGIEE173 pKa = 4.29FVGGDD178 pKa = 3.31VDD180 pKa = 6.46GRR182 pKa = 11.84DD183 pKa = 3.34AWCDD187 pKa = 3.17QSGIRR192 pKa = 11.84SGDD195 pKa = 3.6PLIGTSVRR203 pKa = 11.84SPRR206 pKa = 11.84LPSSGTIGGVCLGLAPQGGPYY227 pKa = 9.23TPPDD231 pKa = 3.56GAEE234 pKa = 4.08TSLSRR239 pKa = 11.84CNANCRR245 pKa = 3.61
Molecular weight: 27.02 kDa
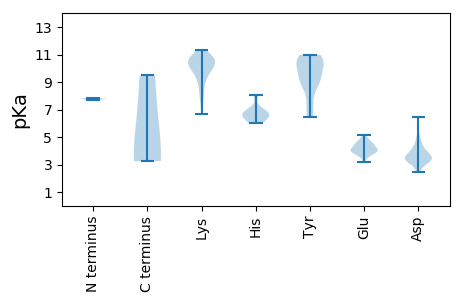
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161IVT0|A0A161IVT0_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 22 OX=1843742 PE=3 SV=1
MM1 pKa = 7.7ARR3 pKa = 11.84LRR5 pKa = 11.84YY6 pKa = 9.11VKK8 pKa = 9.88RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 9.1RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.32PNSRR18 pKa = 11.84RR19 pKa = 11.84STAKK23 pKa = 8.47TRR25 pKa = 11.84PYY27 pKa = 10.02RR28 pKa = 11.84KK29 pKa = 9.32RR30 pKa = 11.84RR31 pKa = 11.84VTPRR35 pKa = 11.84RR36 pKa = 11.84LTKK39 pKa = 10.37KK40 pKa = 10.23AILNITSKK48 pKa = 10.7KK49 pKa = 9.74KK50 pKa = 10.0RR51 pKa = 11.84NTMLSVTNTNSAGVPVPIALGGVFINGTGSGKK83 pKa = 10.38IIWSPTAMDD92 pKa = 5.59LSTDD96 pKa = 4.68DD97 pKa = 4.31IDD99 pKa = 3.71NTAISSSLRR108 pKa = 11.84TSQVCYY114 pKa = 8.69MRR116 pKa = 11.84GVAEE120 pKa = 4.39HH121 pKa = 6.72LRR123 pKa = 11.84IQTSNGLAWFHH134 pKa = 6.55RR135 pKa = 11.84RR136 pKa = 11.84ICFTSKK142 pKa = 10.61APEE145 pKa = 3.94FRR147 pKa = 11.84YY148 pKa = 9.04MAGDD152 pKa = 3.84SAPNAQWNPYY162 pKa = 9.12IEE164 pKa = 4.48NSNGLQRR171 pKa = 11.84FMLNMQVNNMNNTISRR187 pKa = 11.84MEE189 pKa = 3.84TLIFKK194 pKa = 8.93GQQGKK199 pKa = 9.53DD200 pKa = 2.81WSDD203 pKa = 3.5PLIAPLDD210 pKa = 3.89RR211 pKa = 11.84YY212 pKa = 10.66QIDD215 pKa = 3.57VKK217 pKa = 10.79FDD219 pKa = 3.38KK220 pKa = 10.38TWTIRR225 pKa = 11.84SGNEE229 pKa = 3.2KK230 pKa = 9.36GTVAEE235 pKa = 4.21RR236 pKa = 11.84KK237 pKa = 9.0LWHH240 pKa = 6.76PMNKK244 pKa = 9.53NLVYY248 pKa = 10.77ADD250 pKa = 4.77DD251 pKa = 4.19EE252 pKa = 4.71NGTDD256 pKa = 3.31EE257 pKa = 4.48VTSHH261 pKa = 7.05FSVTDD266 pKa = 3.55KK267 pKa = 11.33QGMGDD272 pKa = 3.62YY273 pKa = 10.36YY274 pKa = 10.19IYY276 pKa = 10.54DD277 pKa = 3.43IFIPGAGSSSGDD289 pKa = 3.72YY290 pKa = 10.98INITPSTTIFWHH302 pKa = 6.17EE303 pKa = 4.0KK304 pKa = 9.48
MM1 pKa = 7.7ARR3 pKa = 11.84LRR5 pKa = 11.84YY6 pKa = 9.11VKK8 pKa = 9.88RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 9.1RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.32PNSRR18 pKa = 11.84RR19 pKa = 11.84STAKK23 pKa = 8.47TRR25 pKa = 11.84PYY27 pKa = 10.02RR28 pKa = 11.84KK29 pKa = 9.32RR30 pKa = 11.84RR31 pKa = 11.84VTPRR35 pKa = 11.84RR36 pKa = 11.84LTKK39 pKa = 10.37KK40 pKa = 10.23AILNITSKK48 pKa = 10.7KK49 pKa = 9.74KK50 pKa = 10.0RR51 pKa = 11.84NTMLSVTNTNSAGVPVPIALGGVFINGTGSGKK83 pKa = 10.38IIWSPTAMDD92 pKa = 5.59LSTDD96 pKa = 4.68DD97 pKa = 4.31IDD99 pKa = 3.71NTAISSSLRR108 pKa = 11.84TSQVCYY114 pKa = 8.69MRR116 pKa = 11.84GVAEE120 pKa = 4.39HH121 pKa = 6.72LRR123 pKa = 11.84IQTSNGLAWFHH134 pKa = 6.55RR135 pKa = 11.84RR136 pKa = 11.84ICFTSKK142 pKa = 10.61APEE145 pKa = 3.94FRR147 pKa = 11.84YY148 pKa = 9.04MAGDD152 pKa = 3.84SAPNAQWNPYY162 pKa = 9.12IEE164 pKa = 4.48NSNGLQRR171 pKa = 11.84FMLNMQVNNMNNTISRR187 pKa = 11.84MEE189 pKa = 3.84TLIFKK194 pKa = 8.93GQQGKK199 pKa = 9.53DD200 pKa = 2.81WSDD203 pKa = 3.5PLIAPLDD210 pKa = 3.89RR211 pKa = 11.84YY212 pKa = 10.66QIDD215 pKa = 3.57VKK217 pKa = 10.79FDD219 pKa = 3.38KK220 pKa = 10.38TWTIRR225 pKa = 11.84SGNEE229 pKa = 3.2KK230 pKa = 9.36GTVAEE235 pKa = 4.21RR236 pKa = 11.84KK237 pKa = 9.0LWHH240 pKa = 6.76PMNKK244 pKa = 9.53NLVYY248 pKa = 10.77ADD250 pKa = 4.77DD251 pKa = 4.19EE252 pKa = 4.71NGTDD256 pKa = 3.31EE257 pKa = 4.48VTSHH261 pKa = 7.05FSVTDD266 pKa = 3.55KK267 pKa = 11.33QGMGDD272 pKa = 3.62YY273 pKa = 10.36YY274 pKa = 10.19IYY276 pKa = 10.54DD277 pKa = 3.43IFIPGAGSSSGDD289 pKa = 3.72YY290 pKa = 10.98INITPSTTIFWHH302 pKa = 6.17EE303 pKa = 4.0KK304 pKa = 9.48
Molecular weight: 34.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
877 |
245 |
328 |
292.3 |
32.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.526 ± 0.872 | 2.166 ± 0.75 |
7.526 ± 0.864 | 4.333 ± 0.65 |
4.333 ± 0.489 | 9.464 ± 1.417 |
2.737 ± 0.517 | 6.157 ± 0.501 |
4.789 ± 0.92 | 5.473 ± 0.149 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.052 ± 0.728 | 3.877 ± 1.501 |
5.473 ± 0.735 | 1.938 ± 0.463 |
8.438 ± 0.096 | 6.043 ± 0.982 |
5.359 ± 1.429 | 5.701 ± 0.754 |
2.737 ± 0.289 | 3.877 ± 0.079 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
