
Cellulophaga phage phi47:1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
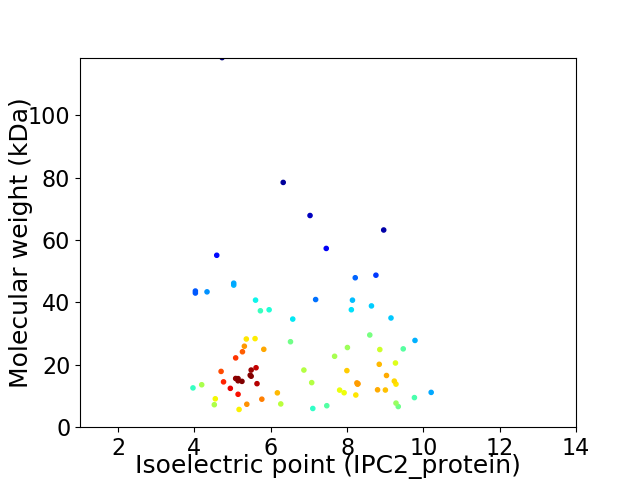
Virtual 2D-PAGE plot for 79 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1PXS0|M1PXS0_9CAUD Structural protein OS=Cellulophaga phage phi47:1 OX=756281 GN=CDPG_00063 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.03NYY4 pKa = 10.66LSILFLLSSFLVLSQNPTSFRR25 pKa = 11.84NGVIPGVKK33 pKa = 7.19TTAEE37 pKa = 3.75IQAIVSPKK45 pKa = 10.56LGTQIFNSTTEE56 pKa = 4.5TIWVRR61 pKa = 11.84QSSSWADD68 pKa = 3.22TGVGGTVTGDD78 pKa = 3.14VVTFDD83 pKa = 3.39SSKK86 pKa = 11.29ANAVDD91 pKa = 3.65VEE93 pKa = 4.53IVDD96 pKa = 4.24GDD98 pKa = 3.81PSVDD102 pKa = 3.35YY103 pKa = 11.1KK104 pKa = 11.56LLFDD108 pKa = 5.0INGTGWGAEE117 pKa = 4.44TPTSTWGQIQGLLSDD132 pKa = 3.85QTDD135 pKa = 3.32LATALNNKK143 pKa = 9.38ISFSGANLGTSTFILGGSGNNIIAFSRR170 pKa = 11.84LDD172 pKa = 3.52NNTTYY177 pKa = 11.3VMSKK181 pKa = 10.16LKK183 pKa = 10.89HH184 pKa = 5.66EE185 pKa = 4.69FSLGNDD191 pKa = 3.72DD192 pKa = 5.8DD193 pKa = 5.27SVTSVYY199 pKa = 10.69TMEE202 pKa = 5.1PDD204 pKa = 4.2KK205 pKa = 10.85FTTDD209 pKa = 3.02LTLAEE214 pKa = 4.48VKK216 pKa = 10.66AGGSGQFLTRR226 pKa = 11.84EE227 pKa = 4.34AGDD230 pKa = 4.27DD231 pKa = 3.75LYY233 pKa = 11.6APAGSTGGSEE243 pKa = 4.33EE244 pKa = 4.06NTDD247 pKa = 3.8NQTLSYY253 pKa = 10.6NDD255 pKa = 3.56YY256 pKa = 11.39GEE258 pKa = 4.11ISITGGNTITATRR271 pKa = 11.84YY272 pKa = 10.08RR273 pKa = 11.84NITTFDD279 pKa = 4.78LIDD282 pKa = 4.72DD283 pKa = 4.72GGGATYY289 pKa = 10.67SPTAFGVSEE298 pKa = 4.27TYY300 pKa = 10.32EE301 pKa = 3.85FSDD304 pKa = 3.54NVLGAVYY311 pKa = 10.19VNPIVTGTPTGDD323 pKa = 3.21LVFDD327 pKa = 4.75LGASYY332 pKa = 10.85EE333 pKa = 4.26VGSTNWSIFDD343 pKa = 3.61VFISNVNNTGDD354 pKa = 3.8DD355 pKa = 4.24YY356 pKa = 11.57YY357 pKa = 11.72LKK359 pKa = 10.45AIKK362 pKa = 8.75RR363 pKa = 11.84TKK365 pKa = 10.35VSSPDD370 pKa = 3.86DD371 pKa = 3.31IYY373 pKa = 11.52KK374 pKa = 10.37IMSVDD379 pKa = 3.25ASGKK383 pKa = 9.61LVPFPAPLSFDD394 pKa = 3.94TINLKK399 pKa = 10.1LQYY402 pKa = 10.8NFYY405 pKa = 10.43PYY407 pKa = 11.3
MM1 pKa = 7.49KK2 pKa = 10.03NYY4 pKa = 10.66LSILFLLSSFLVLSQNPTSFRR25 pKa = 11.84NGVIPGVKK33 pKa = 7.19TTAEE37 pKa = 3.75IQAIVSPKK45 pKa = 10.56LGTQIFNSTTEE56 pKa = 4.5TIWVRR61 pKa = 11.84QSSSWADD68 pKa = 3.22TGVGGTVTGDD78 pKa = 3.14VVTFDD83 pKa = 3.39SSKK86 pKa = 11.29ANAVDD91 pKa = 3.65VEE93 pKa = 4.53IVDD96 pKa = 4.24GDD98 pKa = 3.81PSVDD102 pKa = 3.35YY103 pKa = 11.1KK104 pKa = 11.56LLFDD108 pKa = 5.0INGTGWGAEE117 pKa = 4.44TPTSTWGQIQGLLSDD132 pKa = 3.85QTDD135 pKa = 3.32LATALNNKK143 pKa = 9.38ISFSGANLGTSTFILGGSGNNIIAFSRR170 pKa = 11.84LDD172 pKa = 3.52NNTTYY177 pKa = 11.3VMSKK181 pKa = 10.16LKK183 pKa = 10.89HH184 pKa = 5.66EE185 pKa = 4.69FSLGNDD191 pKa = 3.72DD192 pKa = 5.8DD193 pKa = 5.27SVTSVYY199 pKa = 10.69TMEE202 pKa = 5.1PDD204 pKa = 4.2KK205 pKa = 10.85FTTDD209 pKa = 3.02LTLAEE214 pKa = 4.48VKK216 pKa = 10.66AGGSGQFLTRR226 pKa = 11.84EE227 pKa = 4.34AGDD230 pKa = 4.27DD231 pKa = 3.75LYY233 pKa = 11.6APAGSTGGSEE243 pKa = 4.33EE244 pKa = 4.06NTDD247 pKa = 3.8NQTLSYY253 pKa = 10.6NDD255 pKa = 3.56YY256 pKa = 11.39GEE258 pKa = 4.11ISITGGNTITATRR271 pKa = 11.84YY272 pKa = 10.08RR273 pKa = 11.84NITTFDD279 pKa = 4.78LIDD282 pKa = 4.72DD283 pKa = 4.72GGGATYY289 pKa = 10.67SPTAFGVSEE298 pKa = 4.27TYY300 pKa = 10.32EE301 pKa = 3.85FSDD304 pKa = 3.54NVLGAVYY311 pKa = 10.19VNPIVTGTPTGDD323 pKa = 3.21LVFDD327 pKa = 4.75LGASYY332 pKa = 10.85EE333 pKa = 4.26VGSTNWSIFDD343 pKa = 3.61VFISNVNNTGDD354 pKa = 3.8DD355 pKa = 4.24YY356 pKa = 11.57YY357 pKa = 11.72LKK359 pKa = 10.45AIKK362 pKa = 8.75RR363 pKa = 11.84TKK365 pKa = 10.35VSSPDD370 pKa = 3.86DD371 pKa = 3.31IYY373 pKa = 11.52KK374 pKa = 10.37IMSVDD379 pKa = 3.25ASGKK383 pKa = 9.61LVPFPAPLSFDD394 pKa = 3.94TINLKK399 pKa = 10.1LQYY402 pKa = 10.8NFYY405 pKa = 10.43PYY407 pKa = 11.3
Molecular weight: 43.67 kDa
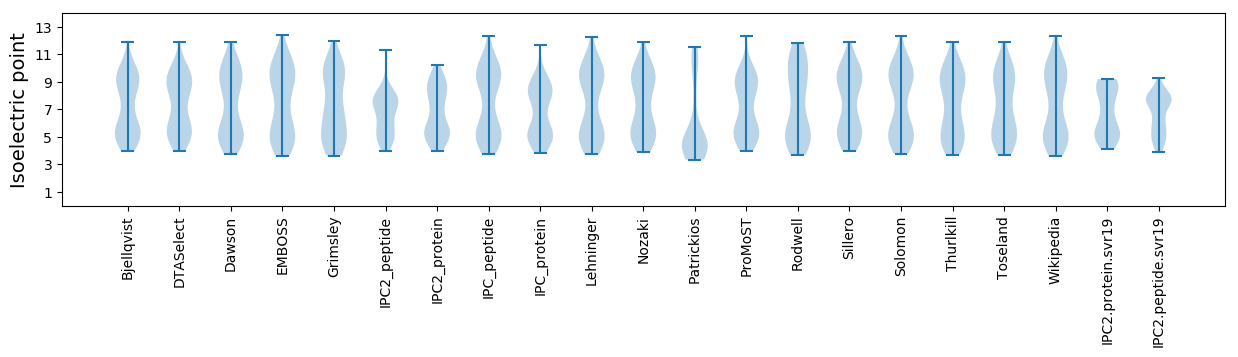
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1Q6Z4|M1Q6Z4_9CAUD Uncharacterized protein OS=Cellulophaga phage phi47:1 OX=756281 GN=CDPG_00024 PE=4 SV=1
MM1 pKa = 6.98YY2 pKa = 10.26KK3 pKa = 10.28SRR5 pKa = 11.84IKK7 pKa = 10.22IQRR10 pKa = 11.84KK11 pKa = 5.85TSYY14 pKa = 11.49SNLEE18 pKa = 3.89PKK20 pKa = 10.09NRR22 pKa = 11.84SIMEE26 pKa = 4.31HH27 pKa = 6.25EE28 pKa = 4.31NQATRR33 pKa = 11.84AMRR36 pKa = 11.84NFSDD40 pKa = 3.6QDD42 pKa = 3.52SMNNSKK48 pKa = 10.96AKK50 pKa = 9.28TISGITAAKK59 pKa = 8.22TASKK63 pKa = 9.76WLRR66 pKa = 11.84NHH68 pKa = 6.39KK69 pKa = 8.22QTNNGKK75 pKa = 9.09NFAKK79 pKa = 10.75NEE81 pKa = 3.95NN82 pKa = 3.69
MM1 pKa = 6.98YY2 pKa = 10.26KK3 pKa = 10.28SRR5 pKa = 11.84IKK7 pKa = 10.22IQRR10 pKa = 11.84KK11 pKa = 5.85TSYY14 pKa = 11.49SNLEE18 pKa = 3.89PKK20 pKa = 10.09NRR22 pKa = 11.84SIMEE26 pKa = 4.31HH27 pKa = 6.25EE28 pKa = 4.31NQATRR33 pKa = 11.84AMRR36 pKa = 11.84NFSDD40 pKa = 3.6QDD42 pKa = 3.52SMNNSKK48 pKa = 10.96AKK50 pKa = 9.28TISGITAAKK59 pKa = 8.22TASKK63 pKa = 9.76WLRR66 pKa = 11.84NHH68 pKa = 6.39KK69 pKa = 8.22QTNNGKK75 pKa = 9.09NFAKK79 pKa = 10.75NEE81 pKa = 3.95NN82 pKa = 3.69
Molecular weight: 9.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17824 |
51 |
1094 |
225.6 |
25.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.559 ± 0.389 | 0.814 ± 0.096 |
6.003 ± 0.198 | 7.058 ± 0.365 |
4.623 ± 0.199 | 6.048 ± 0.323 |
1.565 ± 0.122 | 6.777 ± 0.22 |
9.403 ± 0.649 | 8.259 ± 0.243 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.009 ± 0.137 | 6.413 ± 0.243 |
2.8 ± 0.147 | 3.282 ± 0.138 |
3.944 ± 0.363 | 7.787 ± 0.302 |
5.846 ± 0.253 | 6.351 ± 0.227 |
1.133 ± 0.126 | 3.327 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
