
Alkaliphilus metalliredigens (strain QYMF)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Alkaliphilus; Alkaliphilus metalliredigens
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
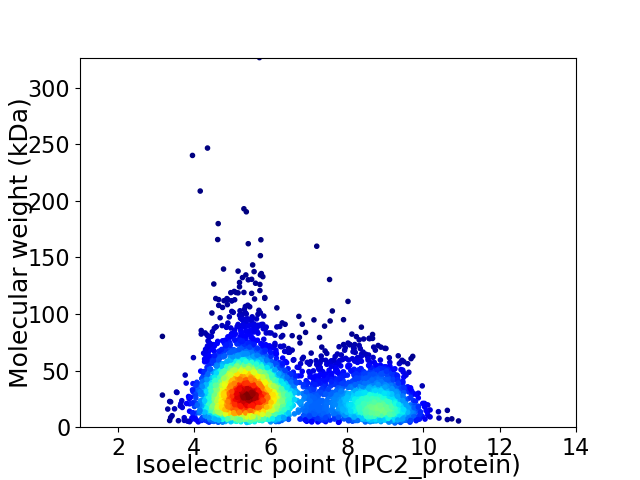
Virtual 2D-PAGE plot for 4467 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6TSE0|A6TSE0_ALKMQ [Citrate [pro-3S]-lyase] ligase OS=Alkaliphilus metalliredigens (strain QYMF) OX=293826 GN=Amet_2958 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.12KK3 pKa = 10.71GLIITLLITSMLFMVGFAPTAGQSYY28 pKa = 7.83WEE30 pKa = 4.21EE31 pKa = 3.83MKK33 pKa = 10.55KK34 pKa = 10.48VYY36 pKa = 9.8EE37 pKa = 3.96WDD39 pKa = 3.36AMEE42 pKa = 5.58GDD44 pKa = 3.68TDD46 pKa = 4.5LEE48 pKa = 4.57VIISVPGQLEE58 pKa = 3.77ATYY61 pKa = 10.43NITMYY66 pKa = 10.81SQTDD70 pKa = 3.62LEE72 pKa = 4.69AFVSYY77 pKa = 10.91IEE79 pKa = 4.18MEE81 pKa = 4.3VEE83 pKa = 3.57IEE85 pKa = 4.03EE86 pKa = 4.48ADD88 pKa = 3.82EE89 pKa = 4.28TTHH92 pKa = 7.03QIPTVKK98 pKa = 9.83MYY100 pKa = 10.55TKK102 pKa = 10.69GSNIYY107 pKa = 10.2INTDD111 pKa = 3.52LVVHH115 pKa = 6.55LLSLAEE121 pKa = 4.11SEE123 pKa = 5.31LPFEE127 pKa = 4.59TEE129 pKa = 3.47AEE131 pKa = 4.29YY132 pKa = 10.95IMLQSGEE139 pKa = 4.5NNMNMDD145 pKa = 4.61LTSLTDD151 pKa = 3.76IIGFLEE157 pKa = 4.25NMDD160 pKa = 4.29LGIDD164 pKa = 3.69LGMTQEE170 pKa = 4.12GDD172 pKa = 3.59TYY174 pKa = 10.75SLRR177 pKa = 11.84IEE179 pKa = 4.11SDD181 pKa = 3.02KK182 pKa = 11.15MIDD185 pKa = 4.15LLDD188 pKa = 3.68VYY190 pKa = 10.45IKK192 pKa = 10.77YY193 pKa = 10.8VITNMDD199 pKa = 3.24QLTNLFMQGDD209 pKa = 4.13VPEE212 pKa = 4.37EE213 pKa = 3.79MVLTEE218 pKa = 3.96EE219 pKa = 4.38QQAEE223 pKa = 4.31MLEE226 pKa = 4.17MYY228 pKa = 10.38EE229 pKa = 4.94MYY231 pKa = 9.1ITPQKK236 pKa = 11.36DD237 pKa = 3.12MAKK240 pKa = 10.32GFILGSYY247 pKa = 8.53YY248 pKa = 10.88DD249 pKa = 3.58QVTTFHH255 pKa = 6.61EE256 pKa = 4.04NSYY259 pKa = 9.84TEE261 pKa = 4.13DD262 pKa = 3.54AEE264 pKa = 5.33LFFTTPMGKK273 pKa = 7.94MHH275 pKa = 7.44IDD277 pKa = 3.97MISSGQKK284 pKa = 10.24LDD286 pKa = 3.32TTTIEE291 pKa = 4.33LPTSVMVITEE301 pKa = 4.34DD302 pKa = 3.39EE303 pKa = 4.48LTNLMISGFGATQDD317 pKa = 3.64MPEE320 pKa = 4.35GSQLQAIVDD329 pKa = 4.03LQGDD333 pKa = 3.92YY334 pKa = 10.92MKK336 pKa = 9.62ITEE339 pKa = 4.2GEE341 pKa = 4.2VLEE344 pKa = 4.81GNINLKK350 pKa = 10.21VEE352 pKa = 4.29EE353 pKa = 4.42GRR355 pKa = 11.84SYY357 pKa = 11.04IVVEE361 pKa = 5.13DD362 pKa = 3.63IQEE365 pKa = 4.13LLDD368 pKa = 3.75IEE370 pKa = 4.71FEE372 pKa = 4.13GLEE375 pKa = 4.29GFMMIQDD382 pKa = 4.43LEE384 pKa = 4.33QYY386 pKa = 10.55GYY388 pKa = 10.44QIQWNPDD395 pKa = 2.61SRR397 pKa = 11.84TIEE400 pKa = 3.91IYY402 pKa = 10.7NN403 pKa = 3.43
MM1 pKa = 7.62KK2 pKa = 10.12KK3 pKa = 10.71GLIITLLITSMLFMVGFAPTAGQSYY28 pKa = 7.83WEE30 pKa = 4.21EE31 pKa = 3.83MKK33 pKa = 10.55KK34 pKa = 10.48VYY36 pKa = 9.8EE37 pKa = 3.96WDD39 pKa = 3.36AMEE42 pKa = 5.58GDD44 pKa = 3.68TDD46 pKa = 4.5LEE48 pKa = 4.57VIISVPGQLEE58 pKa = 3.77ATYY61 pKa = 10.43NITMYY66 pKa = 10.81SQTDD70 pKa = 3.62LEE72 pKa = 4.69AFVSYY77 pKa = 10.91IEE79 pKa = 4.18MEE81 pKa = 4.3VEE83 pKa = 3.57IEE85 pKa = 4.03EE86 pKa = 4.48ADD88 pKa = 3.82EE89 pKa = 4.28TTHH92 pKa = 7.03QIPTVKK98 pKa = 9.83MYY100 pKa = 10.55TKK102 pKa = 10.69GSNIYY107 pKa = 10.2INTDD111 pKa = 3.52LVVHH115 pKa = 6.55LLSLAEE121 pKa = 4.11SEE123 pKa = 5.31LPFEE127 pKa = 4.59TEE129 pKa = 3.47AEE131 pKa = 4.29YY132 pKa = 10.95IMLQSGEE139 pKa = 4.5NNMNMDD145 pKa = 4.61LTSLTDD151 pKa = 3.76IIGFLEE157 pKa = 4.25NMDD160 pKa = 4.29LGIDD164 pKa = 3.69LGMTQEE170 pKa = 4.12GDD172 pKa = 3.59TYY174 pKa = 10.75SLRR177 pKa = 11.84IEE179 pKa = 4.11SDD181 pKa = 3.02KK182 pKa = 11.15MIDD185 pKa = 4.15LLDD188 pKa = 3.68VYY190 pKa = 10.45IKK192 pKa = 10.77YY193 pKa = 10.8VITNMDD199 pKa = 3.24QLTNLFMQGDD209 pKa = 4.13VPEE212 pKa = 4.37EE213 pKa = 3.79MVLTEE218 pKa = 3.96EE219 pKa = 4.38QQAEE223 pKa = 4.31MLEE226 pKa = 4.17MYY228 pKa = 10.38EE229 pKa = 4.94MYY231 pKa = 9.1ITPQKK236 pKa = 11.36DD237 pKa = 3.12MAKK240 pKa = 10.32GFILGSYY247 pKa = 8.53YY248 pKa = 10.88DD249 pKa = 3.58QVTTFHH255 pKa = 6.61EE256 pKa = 4.04NSYY259 pKa = 9.84TEE261 pKa = 4.13DD262 pKa = 3.54AEE264 pKa = 5.33LFFTTPMGKK273 pKa = 7.94MHH275 pKa = 7.44IDD277 pKa = 3.97MISSGQKK284 pKa = 10.24LDD286 pKa = 3.32TTTIEE291 pKa = 4.33LPTSVMVITEE301 pKa = 4.34DD302 pKa = 3.39EE303 pKa = 4.48LTNLMISGFGATQDD317 pKa = 3.64MPEE320 pKa = 4.35GSQLQAIVDD329 pKa = 4.03LQGDD333 pKa = 3.92YY334 pKa = 10.92MKK336 pKa = 9.62ITEE339 pKa = 4.2GEE341 pKa = 4.2VLEE344 pKa = 4.81GNINLKK350 pKa = 10.21VEE352 pKa = 4.29EE353 pKa = 4.42GRR355 pKa = 11.84SYY357 pKa = 11.04IVVEE361 pKa = 5.13DD362 pKa = 3.63IQEE365 pKa = 4.13LLDD368 pKa = 3.75IEE370 pKa = 4.71FEE372 pKa = 4.13GLEE375 pKa = 4.29GFMMIQDD382 pKa = 4.43LEE384 pKa = 4.33QYY386 pKa = 10.55GYY388 pKa = 10.44QIQWNPDD395 pKa = 2.61SRR397 pKa = 11.84TIEE400 pKa = 3.91IYY402 pKa = 10.7NN403 pKa = 3.43
Molecular weight: 46.04 kDa
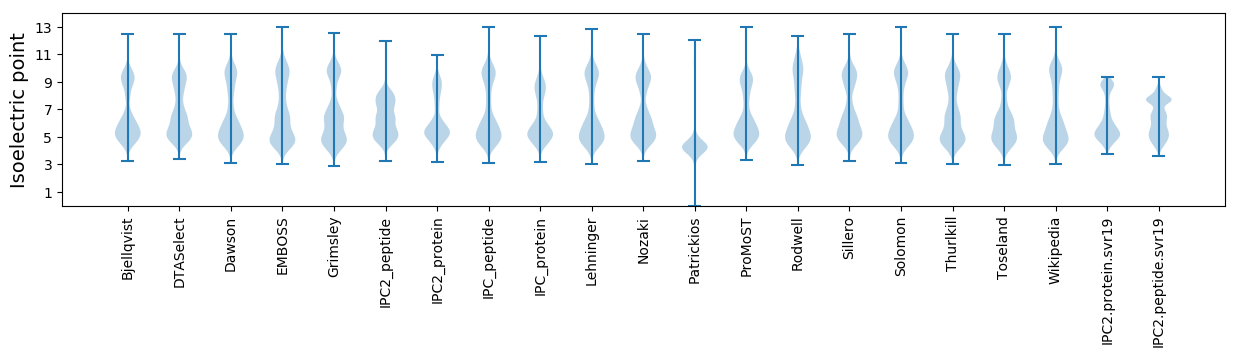
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6TJG9|A6TJG9_ALKMQ Nicotinate phosphoribosyltransferase OS=Alkaliphilus metalliredigens (strain QYMF) OX=293826 GN=Amet_0094 PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.0KK9 pKa = 7.87RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.47EE15 pKa = 3.72HH16 pKa = 6.09GFVKK20 pKa = 10.46RR21 pKa = 11.84MRR23 pKa = 11.84TTSGRR28 pKa = 11.84RR29 pKa = 11.84ILKK32 pKa = 9.77RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.28GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.0KK9 pKa = 7.87RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.47EE15 pKa = 3.72HH16 pKa = 6.09GFVKK20 pKa = 10.46RR21 pKa = 11.84MRR23 pKa = 11.84TTSGRR28 pKa = 11.84RR29 pKa = 11.84ILKK32 pKa = 9.77RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.28GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1321948 |
34 |
2843 |
295.9 |
33.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.331 ± 0.035 | 0.924 ± 0.014 |
5.189 ± 0.027 | 7.81 ± 0.044 |
4.125 ± 0.032 | 6.997 ± 0.04 |
1.893 ± 0.016 | 9.05 ± 0.034 |
7.314 ± 0.04 | 9.439 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.019 ± 0.018 | 4.82 ± 0.027 |
3.211 ± 0.022 | 3.491 ± 0.026 |
4.021 ± 0.023 | 5.803 ± 0.029 |
5.214 ± 0.029 | 6.893 ± 0.029 |
0.782 ± 0.012 | 3.676 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
